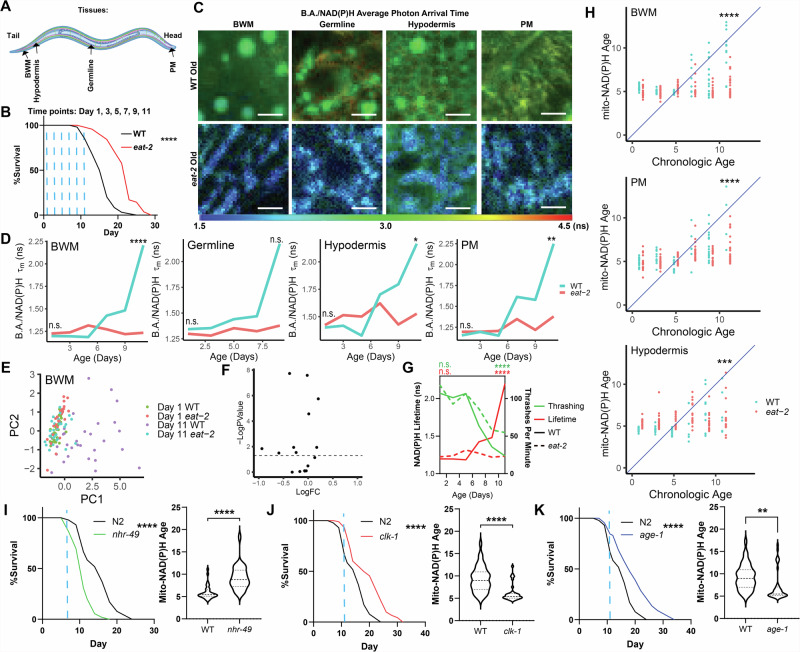
Fig. 4. Mitochondrial B.A./NAD(P)H FLIM endpoints are linked to lifespan.
A,B Schematic of imaging locations throughout the C. elegans body (A) and time points (B; blue dashed lines) imaged for data presented in Fig. 4. B shows a lifespan curve for WT (N2) and eat-2 (ad1116) C. elegans (n = 100 C. elegans per condition, p < 0.0001, Log-rank (Mantel–Cox) test). C,D WT (blue) and eat-2 (red) B.A./NAD(P)H average photon arrival time images and analysis of C. elegans in the germline, PM, BWM, and hypodermis throughout age (n = 5–32 C. elegans per condition; moderated t test; median). E PCA plots of non-redundant B.A./NAD(P)H FLIM endpoints (α1, Ƭ1, Ƭ2 and intensity) of Day 1 and Day 11 WT and eat-2 C. elegans BWM mitochondria B.A./NAD(P)H FLIM endpoints. Each dot is an individual C. elegans (n = 24–32 C. elegans per condition). F Volcano plot summarizing statistical analyzes of B.A./NAD(P)H FLIM endpoints comparing Day 11 WT and eat-2 C. elegans to each other. (moderated t test). G Plot of thrashing rate (green) and BWM B.A./NAD(P)H lifetime (red) in WT (solid line) or eat-2 (dashed line) C. elegans versus age (n = 21–32 C. elegans per condition; Two-way ANOVA with post-hoc Tukey’s test; median (B.A./NAD(P)H FLIM), mean (thrashing)). H Mito-NAD(P)H Age for WT (blue) and eat-2 (red) animals using LASSO regression models generated in Figs. 1 and 3 plotted versus age. Each dot represents an individual C. elegans (n = 13–32 C. elegans per condition; Student’s t test). I–K Lifespan and mitochondrial BWM Mito-NAD(P)H Age in WT, nhr-49, clk-1 and age-1 mutants at the indicated time points (blue dashed line – Day 7 for nhr-49 or Day 11 for age-1 and clk-1; n = 100 C. elegans per condition in lifespans and n = 27–40 for B.A./NAD(P)H FLIM analyzes, Log-rank (Mantel–Cox) test (lifespans) Student’s t test (mito-NAD(P)H Age). Scale bars, 10 µm. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.