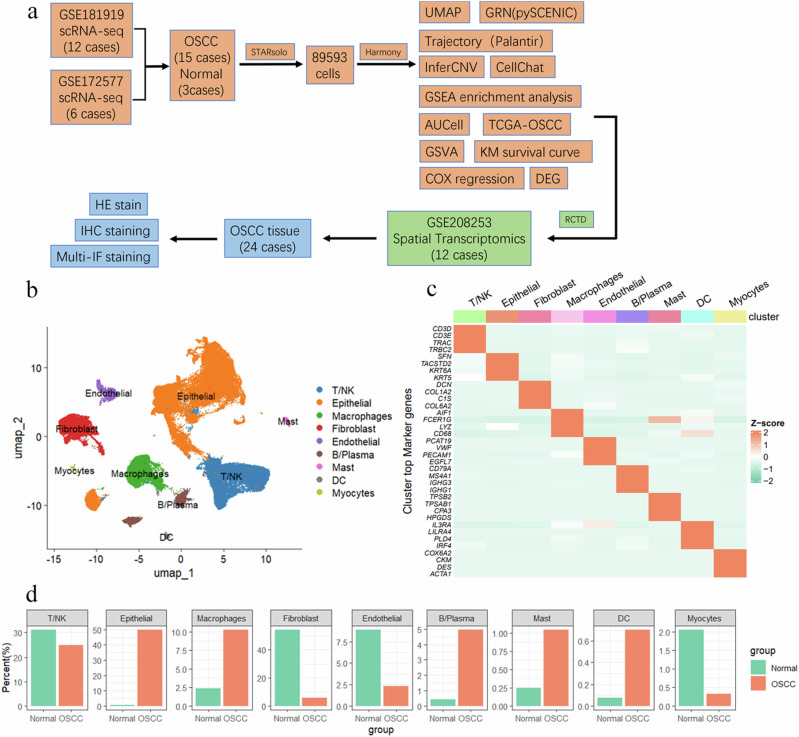
Fig. 1. Integration of OSCC scRNA-seq Atlas.
a The workflow diagram illustrates the data analysis process. The data were summarized and categorized into 15 OSCC samples and 3 normal samples. Initial data processing was conducted using the STARsolo software, resulting in a total of 89,593 cell data points. The Harmony algorithm was applied for batch effect correction. Various analytical methods were employed, including UMAP dimensionality reduction, GRN analysis (using pySCENIC), trajectory analysis (using Palantir), InferCNV, and CellChat. Single-cell annotations were mapped to spatial transcriptomics using RCTD, and further validation was conducted through histological staining. b UMAP plot showing nine major cell types: T/NK cells, epithelial cells, macrophages, fibroblasts, endothelial cells, B/plasma cells, mast cells, DC cells, and myocytes. c Heatmap showing the expression of representative marker genes for each cell type. Cell clusters are indicated on the x-axis, and gene names are on the y-axis. The color intensity represents the average gene expression level, with Z-score values standardized. d Proportion of cell types in OSCC patients; the y-axis represents the percentage of cell counts, and the x-axis represents the Normal and OSCC groups.