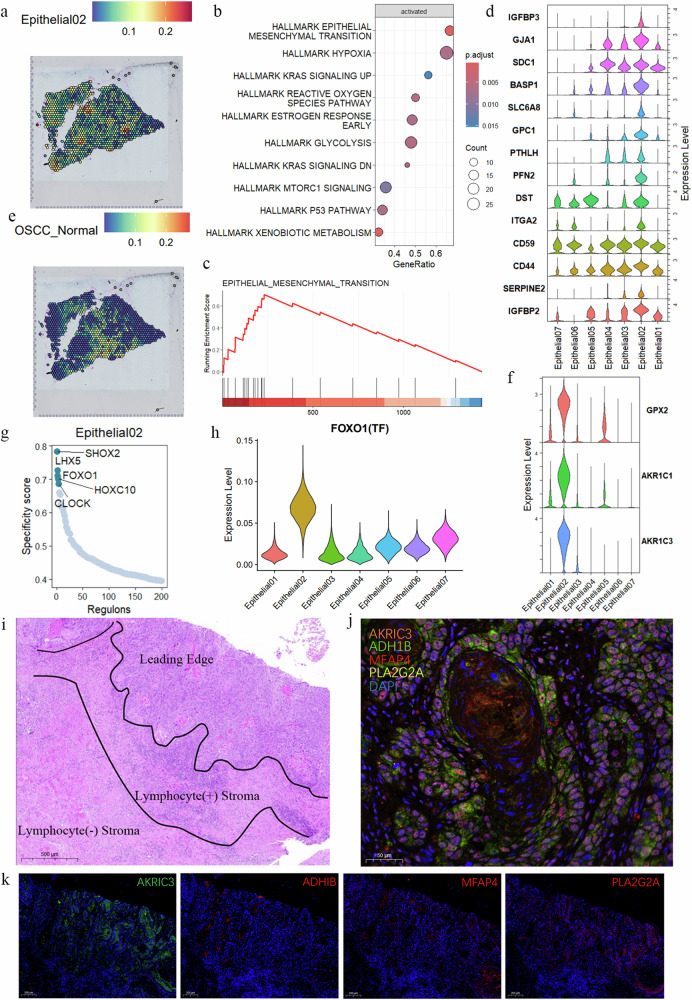
Fig. 7. Evidence of interaction between Epithelial02 tumor cell clusters and OSCC–Normal.
a, e The spatial transcriptomics display the distribution of Epithelial02 and OSCC_Normal in OSCC samples. The top of each panel shows the expression intensity for each cell cluster, with darker colors indicating higher intensity. b GSEA analysis results display the biological processes activated in Epithelial02. c GSEA results show the enrichment scores of EMT in Epithelial02. d The violin plot presents the expression of EMT genes in epithelial cells. f The violin plot shows the expression of GPX2, AKR1C1, and AKR1C3 in epithelial subtype cells. g Specific transcriptional regulators in Epithelial02. h The violin plot illustrates the expression of transcription factor FOXO1 in epithelial subtype cells. i HE-stained tumor sections, including the invasive front, lymphocyte-positive stromal region, and lymphocyte-negative stromal region. j, k Immunofluorescence staining highlights the specific genes in Epithelial02 and OSCC_Normal (AKR1C3, MFAP4, PLA2G2A, and ADH1B). The nuclei of the cells were stained blue with DAPI.