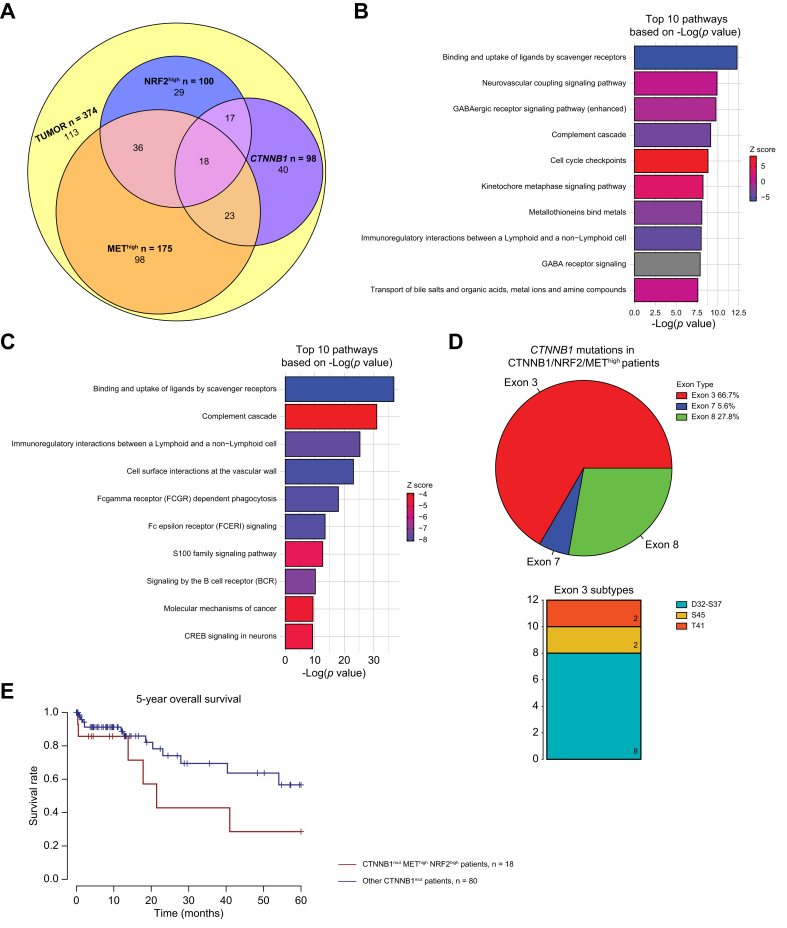
Fig. 1.
Influence of Nrf2 and Met pathway activation on gene expression in HCC with and without CTNNB1-mutations.
(A) Venn diagram of 374 The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) patients categorized as Nrf2-high, Met-high, or CTNNB1-mutated, and their patient overlap. Eighteen (4.8% all HCC) patients had overlap of CTNNB1-mutation, Nrf2-high, Met-high. (B) Top 10 pathways based on p value from Ingenuity pathway analysis (IPA) of differentially expressed genes comparing patients categorized as Nrf2-/Met-high (n = 54) vs. normal (n = 50). Specifically, 5,238 differentially expressed genes were applied to IPA analysis with cutoff of false discovery rate (FDR) = 0.001 and absolute logFC >3. (C) Top 10 pathways based on p value from IPA of differentially expressed genes comparing patients categorized as CTNNB1-mutated/Nrf2-/Met-high (n = 18) vs. normal (n = 50). Specifically, 5,114 differentially expressed genes were subjected to IPA analysis with cutoff of FDR = 0.001 and absolute logFC >3. For both (A) and (B) ranking of pathways based on -log(p value) and activation/inhibition of pathway determined by z-score. Percentages and frequencies are shown. (D) (Left) Pie chart depicting the distribution of exon mutations and (Right) stacked bar plot depicting the frequency of different exon 3 mutations in CTNNB1-mutated/Nrf2-/Met-high patients. (E) Kaplan-Meier curve showing trending decreased overall survival (OS) in CTNNB1-mutated/Nrf2-/Met-high (n = 18) compared with other CTNNB1 mutated cases (n = 80). Log-rank test p = 0.104. Levels of significance: p <0.05, ∗∗p <0.001, ∗∗∗p <0.0001.