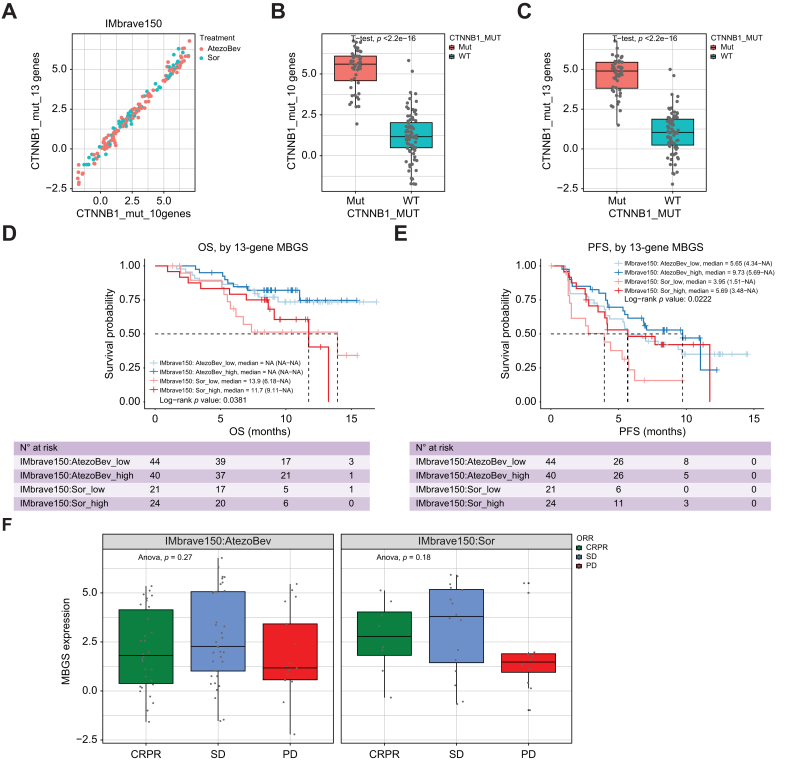
Fig. 7.
MBGS predicts relative response to sorafenib in IMbrave150 trial cohort.
(A) Correlation based on expression of 10-gene and 13-gene mutated-β-catenin gene signature (MBGS) in IMbrave150 trial cohort. (B) Box plot of expression of 10-gene MBGS in CTNNB1 wild-type (n = 82) and mutant (n = 48) cases in IMbrave150 cohort. Student’s t-test p value comparing mutated vs. wild-type patients is ∗∗∗p <2.22e-16. (C) Box plot of expression of 13-gene MBGS in CTNNB1 wild-type (n = 82) and mutant (n = 48) cases in the IMbrave150 cohort. Student’s t-test p value comparing mutated vs. wild-type patients is ∗∗∗p <2.22e-16. For (B) and (C) Individual values per patient are depicted with bold line in middle representing the median and outside boxes showing inner quartile ranges (D) Kaplan–Meier curve for overall survival demonstrating improved response to sorafenib in MBGS-high patients. Log-rank p value is ∗p = 0.0381. (E) Kaplan–Meier curve for progression-free survival (PFS) demonstrating improved response to sorafenib in MBGS-high patients. Response to atezolizumab/bevacizumab is comparable between MBGS-high/low patients. Log-rank p value is ∗p = 0.0222. Log-rank test was used to determine differences in mean survival time. (F) MBGS expression stratified by complete/partial response (CR/PR), stable disease (SD), or progressive disease (PD) defined by mRECIST criteria in each arm. Higher MBGS expression correlated well with sorafenib response. In atezo/bev arm, one-way ANOVA p = 0.27. In sorafenib arm, One-way ANOVA p = 0.18. For (F), individual values per patient are depicted with bold line in middle representing the median and outside boxes showing inner quartile ranges; no statistical test was used but depicted this way for visual representation across the different subclasses. Levels of significance: ∗p <0.05, ∗∗p <0.001, ∗∗∗p <0.0001.