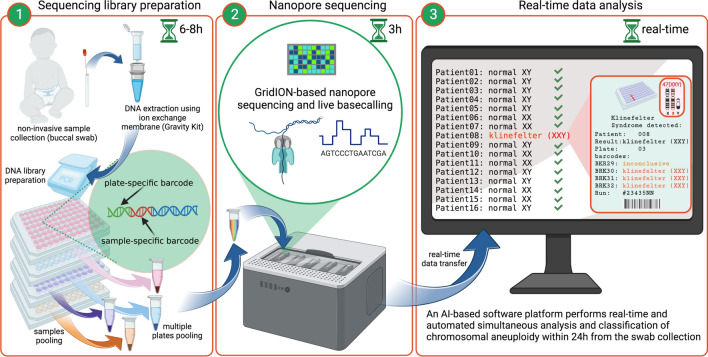
Fig. 1.
Workflow of SCAN. The workflow of the method is composed of three main steps, enabling classification of the five most common chromosomal aneuploidies in less than 24 h. 1. Non-invasive biological material is collected for DNA extraction using a buccal swab. Samples undergo DNA extraction with an ion-exchange membrane, which ensures high molecular weight DNA. Next, samples pass through a two-step PCR protocol that facilitates non-saturated amplification of selected regions representing the chromosomes of interest and reference chromosome 15 (PCR1), followed by sample/technical replicate barcoding (PCR2). Multiple 96-well PCR plates are then pooled within each plate, and each pool receives a plate-specific barcode using the Native Barcoding Kit by Oxford Nanopore Technologies. 2. Sequencing is performed on GridIONx5, where basecalling and first level demultiplexing (plate-specific) occur in real-time. 3. Newly generated data is transferred to the software platform, where automated second-level demultiplexing (by sample/technical replicate) is conducted using Torchlex [24]. Read annotation is performed in real-time, enabling rapid sample classification as soon as the minimum required data is collected