Abstract
Background
Cuproptosis, a metal-ion-dependent form of regulated cell death induced by copper overload, is emerging as a potential mechanism in high-grade glioma (HGG). Despite its significance, the role of cuproptosis in predicting the prognostic and therapeutic response in HGG remains poorly understood.
Methods
We performed unsupervised clustering to stratify patients with HGG in the Chinese Glioma Genome Atlas (CGGA) according to the expression of 14 cuproptosis-related genes (CRGs) and validated in The Cancer Genome Atlas (TCGA). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were applied to explore the biological processes and pathways involved within distinct groups. We constructed the CupScore model to predict the responsiveness to immune checkpoint inhibitors (ICIs) therapy and chemotherapy in patients with HGG. Additionally, in vivo and in vitro experiments were performed to investigate the potential biological function of CDKN2A in HGG.
Results
We identified two cuproptosis-related molecular subgroups with significantly different survival probabilities. Patients with HGG in cluster 1 were characterized as immune-desert phenotype with higher CupScore and lower expression of MHC complex, interferons, chemokines, interleukins, and immune checkpoints. In contrast, cluster 2 showed an immune-inflamed signature. We screened PI-103 as the most promising candidate for patients with higher CupScore and confirmed its experimental evidence and clinical trial status. Patients with lower CupScore showed higher response rates to anti-PD-L1 and anti-PD1 combined with anti-CTLA4 ICI therapy. Furthermore, in vivo and in vitro experiments revealed that CDKN2A enhanced the malignant phenotype of HGG.
Conclusion
Cuproptosis has the ability to reprogram the tumor microenvironment (TME) in HGG, leading to the stratification of patients into two distinct molecular subgroups. The CupScore model emerged as a robust metric for predicting the prognostic and therapeutic benefits, as well as may therefore facilitate personalized treatment strategies for patients with HGG.
Keywords: cuproptosis, high-grade glioma, molecular subtype, prognosis prediction, therapeutic response
Introduction
High-grade gliomas (HGGs) are the most common primary central nervous system (CNS) malignancies and account for approximately 50% of all primary brain tumors, representing the leading cause of death among primary CNS tumors.1 According to the 2021 WHO classification of CNS tumors, HGGs are classified into four subtypes: grade 3 oligodendroglioma (1p/19 codeleted, IDH-mutant); grade 3 IDH-mutant astrocytoma; grade 4 IDH-mutant astrocytoma, and grade 4 IDH wild-type glioblastoma (GBM).2 Survival rates for gliomas vary significantly depending on the subtype. Low-grade gliomas (LGG) can have relatively high 5-year survival rates, reaching up to 80%. On the other hand, HGGs have much lower 5-year survival rates, typically below 5%.3 Furthermore, HGGs are characterized by their high infiltrative nature and resistance to therapy, making them largely incurable. Hence, the pursuit of effective therapeutic strategies for HGGs stands as a pivotal endeavor towards enhancing patient survival rates and prognostic outcomes.
Regulated cell death (RCD), encompassing autophagy, apoptosis, pyroptosis, necroptosis, and ferroptosis, plays a crucial role in tumor biology and is linked to cancer metastasis and the remodeling of the tumor microenvironment (TME).4 Metal ion-dependent forms of RCD, particularly ferroptosis, have been implicated in the progression and therapeutic responses of a range of tumors.5 Notably, copper, an essential cofactor for several enzymes, exhibits elevated levels in certain cancer types, including brain tumors,6 contributing to PD-L1 expression modulation in glioma7 and metastasis in triple-negative breast cancer.8
Cuproptosis, an emerging form of RCD triggered by excessive intracellular copper accumulation,9 has been associated with the destabilization of iron-sulfur cluster proteins.10 A recent study identified key regulators of cuproptosis and underscored its potential impact on inflammation-associated immunosuppression and TME reprogramming, particularly in glioma.11 The cuproptosis-related signature has been linked to immune infiltration and patient prognosis in glioma,12 suggesting its significance in HGG cluster identification and prognostic prediction.
Amidst the inter- and intra-tumor heterogeneity of HGG, precise molecular subtype classification becomes imperative for personalized treatment strategies. Previous studies, including The Cancer Genome Atlas (TCGA) classification and various signatures related to m6A regulation,13 immune response,14 autophagy,15 and ferroptosis,16 have shed light on HGG molecular subtyping, but treatment options remain limited and the mortality rates remain high. In this context, exploring the molecular characteristics of cuproptosis-related genes (CRGs) may unravel the heterogeneity of HGG, offering potential avenues for effective classification schemes and precision therapeutic strategies.
The aim of the present study was to delineate the CRG-based molecular subtypes in HGG, and introduce a CupScore model derived from the expression of 14 CRGs. The study further screened potential chemotherapeutic compounds for HGG with a high CupScore, identifying PI-103 as a promising candidate. Additionally, the potential of the CupScore model as a robust metric for predicting immune checkpoint inhibitors (ICIs) therapy benefits was validated, highlighting novel perspectives for the clinical treatment of HGG.
Materials and Methods
Data Acquisition and Filtration
HGG transcriptomic profiles and matched clinical information were obtained from the China Glioma Genome Atlas (CGGA) (http://www.cgga.org.cn/, DataSet ID: mRNAseq_693) and The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/). The Genotype-Tissue Expression (https://www.genome.gov/) brain tissue mRNA-seq data served as a control. Patients lacking matched survival information were excluded, resulting in the inclusion of 236 patients from CGGA, 165 tumors from TCGA, and 207 normal samples.
Cuproptosis-Related Regulator Identification and Expression Profile Characterization
A total of 14 CRGs were identified from the recent publications.17,18 The mRNA expression profiles of these 14 genes were analyzed in patients with HGG using the data obtained from TCGA and CGGA. Protein expression profiles were obtained using immunohistochemistry (IHC) samples downloaded from the Human Protein Atlas database (HPA; https://www.proteinatlas.org/). The genetic variation landscape of these 14 genes in TCGA HGG was visualized using the “maftools” R package.19 We explored the expression profiles of 14 genes at the single-cell level using eight single-cell RNA sequencing datasets obtained from the Tumor Immune Single-cell Hub (http://tisch.comp-genomics.org/home/). We have uploaded all the R codes for to Github (https://github.com/xiaoying-codeLab/Cuproptosis.GBM.git). We investigated subcellular localization using immunocytochemistry (ICC) data from the Human Protein Atlas (HPA) in the human glioblastoma cell line U251.
Unsupervised Clustering Based on Cuproptosis-Related Genes
Unsupervised clustering analysis was performed using the “ConsensusClusterPlus” R package based on the mRNA expression of the 14 CRGs.20 Principal component analysis (PCA) was employed by “factoextra” R package to demonstrate distribution differences,21 and Kaplan-Meier survival analysis was applied by “Survival” R package to compare patient survival between the different groups.22 The HGG data obtained from TCGA was used for unsupervised clustering validation.
Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Variation Analysis (GSVA) pathway enrichment analysis
Differentially expressed genes (DEGs) between cuproptosis subtypes were identified using the “limma” R package.23 “ClusterProfiler” R package was used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and the GSVA algorithm explored the significantly differentially enriched pathways within clusters.24
Construction of the CupScore Model
To evaluate the prognostic value of cuproptosis signatures, we utilized a penalized Cox regression model with LASSO penalties to calculate correlation coefficients for the 14 CRGs. These coefficients, derived from LASSO regression, quantify the relationship between gene expression and patient prognosis. By multiplying the gene expression levels with their respective coefficients, we obtain the CupScore, which sums up the results to represent the combined prognostic significance of the CRGs. Therefore, the CupScore formula is defined as follows: CupScore = ∑ (correlation coefficient × gene expression). A total of 236 cases were stratified into high and low CupScore groups based on the optimal cutoff value determined by “Survminer” R package.25 Subsequently, survival analysis was conducted to compare the patient outcomes between these two groups. Finally, the “survivalROC” R package was used to construct a Receiver Operating Characteristic (ROC) curve, thus assessing the predictive efficacy of the CupScore model.26
Tumor Immune Infiltration Analysis
Dying cancer cells trigger an immune response, impacting the quantity and composition of immune infiltrations to reshape the TME and thereby contributing to apoptosis resistance and immune evasion.27 To assess these effects, the “ESTIMATE” R package was used to predict the stromal, immune, and overall ESTIMATE scores across distinct cuproptosis subtypes in HGG.28 Furthermore, we computed the Tumor Inflammation Signature (TIS) score by evaluating the expression of an 18-gene set,29 which was developed through a cross-validated penalized regression modeling strategy on a large cohort of pembrolizumab-treated patients across nine different tumor types. This score was used to compare the differences in T cell infiltration among the various CupScore clusters.
Correlation Between the CupScore and Drug Sensitivity
The area under the curve (AUC) values of drug responses were compared among clusters to pinpoint effective medications, leveraging the Genomics of Drug Sensitivity in Cancer (GDSC) database, which contains 367 compounds (https://www.cancerrxgene.org/). Expression data for human cancer cell lines (CCLs) were accessible through the Cancer Cell Line Encyclopedia (https://sites.broadinstitute.org/ccle/). The PRISM dataset (https://www.theprismlab.org/) and the Cancer Therapeutics Response Portal (CTRP2.0) were used to identify potential therapeutic agents for patients with high-grade glioma. Subsequently, the Connectivity Map (CMap) score was employed to identify promising compounds for patients with an elevated CupScore. Information regarding target genes, clinical trial status, and experimental evidence of chemotherapeutic agents was sourced from Drugbank (https://go.drugbank.com/) and the National Center for Biotechnology Information (NCBI) (https://pubmed.ncbi.nlm.nih.gov/).
Correlation Between the CupScore and ICI Therapy Response
An urothelial cancer cohort treated with anti-PD-L1 immunotherapy was used to delve deeper into the association between the CupScore and immunotherapeutic benefits, utilizing the “IMvigor210CoreBiologies” R package. The present study included a total of 298 patients with comprehensive clinical information, excluding cases lacking detailed information regarding the effectiveness of immunotherapy. Concurrently, the predictive value of the CupScore in anti-PD-1 and anti-CTLA-4 combination therapy was investigated by incorporating patients with melanoma from PRJEB23709.
Histological Analysis
High-grade glioma and their corresponding normal tissue samples were procured from the Department of Neurosurgery at Huaian Cancer Hospital (Huaian, China) between January 2020 and September 2022. The research protocol was approved by the Ethics Committee of the Huaian Cancer Hospital (No.2019036) and the present study conformed with the guidelines described in the Declaration of Helsinki.30 Informed patient consent was obtained prior to the inclusion of patients. Tissues were preserved in 10% neutral formalin fixative and sectioned into 5 μm-thick slices. These sections underwent a comprehensive staining regimen, using hematoxylin (RT for 3 min) and eosin (RT for 30 sec) (H&E), copper salt (dithiooxamide method; cat. no. G3040; Beijing Solarbio Science & Technology Co., Ltd)., and IHC staining with an anti-SLC31A1 antibody incubated overnight at 4°C (cat. no. 67221-1-Ig, dilution 1:1000, ProteinTech Group, Inc).
Reverse Transcription-Quantitative PCR (RT-qPCR)
Total RNA was extracted from high-grade glioma and paired normal tissues using an RNA isolation kit (cat. no. RC112-01, Vazyme Biotech Co., Ltd)., and 1 μg RNA was used for reverse transcription (cat. no. R323-01, Vazyme Biotech Co., Ltd). Gene expression was normalized to GAPDH expression. The sequences of the primers used were: GAPDH forward, GTCTCCTCTGACTTCAACAGCG and reverse, ACCACCCTGTTGCTGTAGCCA A; SLC31A1 forward, CCAGGACCAAATGGAACCATCC and reverse, ACCACCTGGATGATGTGCAGCA; and CDKN2A forward, CTCGTGCTGATGCTACTGAGGA and reverse, GGTCGGCGCAGTTGGGCTCC.
Cell Culture
Human U87MG ATCC (glioblastoma of unknown origin; #CL-0238) and U251 (#CL-0237) cells verified using short tandem repeat profiling, were obtained from Procell Life Science & Technology Co., Ltd. The 293T cell line was purchased from Shanghai Cafa Biological Technology Co. Ltd (Shanghai, China). Cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc)., supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc)., and incubated in a humidified incubator at 37°C supplied with 5% CO2 air.
shRNA Preparation and CDKN2A Knockdown
The lentiviral shRNA vector (pGLV-U6/Puro), designed to target human CDKN2A, was constructed and manufactured by GemPharmatech; the specific shRNA sequence was GCTCTGAGAAACCTCGGGAAA.
For the lentivirus packaging process, a recombinant lentiviral vector plasmid (pGLV-U6/Puro-sh CDKN2A, 12 μg), a packaging plasmid (psPAX2, 9 μg), and an envelope plasmid (pMD2G, 3 μg) were utilized to transfect 4×106 293T cells in a 10 cm dish. The plasmids were resuspended in 1.5 mL Opti-MEM and then mixed with an additional 1.5 mL Opti-MEM containing 60 μL polyetherimide, and incubated at room temperature for 20 min. The resulting mixture was added dropwise to the 293 cells. After 8 h, the culture media was replaced with fresh media.
Following a 24 and 48 h incubation period, the culture medium, now containing the lentivirus, was collected. Subsequently, U87MG cells were infected with the lentiviruses and subjected to selection using puromycin (2 μg/mL) to establish stable cell lines.
Proliferation Assay
U87MG and U251 cells, transfected with either control or CDKN2A shRNA, were plated in a 48-well dish at a density of 1×104 cells per well in serum-free medium. Subsequently, the cells were labeled using CFDA-SE (cat. no. C1031, Beyotime Institute of Biotechnology).
Gap Closure and Migration Assays
U87MG and U251 cells,31,32 transfected with either control or CDKN2A shRNA, were seeded in two-well culture inserts (cat. no. 80209; Ibidi GmbH) placed within a 24-well cell culture plate. The following day, the culture inserts were carefully removed, and the closure of the resulting cell-free gaps were monitored continuously for 12 h under a microscope.
Subcutaneous Tumor Growth Assay
U87MG cells, stably infected with lenti-shControl or lenti-shCDKN2A (GemPharmatech), were injected at a density of 3×106 cells in 0.1 mL PBS into nude mice aged 6–8 weeks (n=4). The developing tumor was assessed every 2 days to ensure that its maximum size did not exceed 2.0 cm. Tumor dimensions were measured using calipers, and the volume was calculated using the formula: V = (S2 × L)/2, where V is the volume, S is the shortest diameter, and L is the longest diameter. On day 15, the mice were euthanized using 100% CO2 asphyxiation with a chamber volume displacement rate of 50% per min, for a minimum duration of 3 min. Cervical dislocation was then performed to confirm death. Subsequently, the tumors were isolated, imaged, and measured. All animal procedures were approved by the Laboratory Animal Core Facility of Nantong Medical University.
Statistical Analysis
Data analysis was performed using R version 4.0.4. Comparisons between groups were performed using a Wilcoxon test, Student’s t-test, or one-way ANOVA. To evaluate the equality of variances among the different groups, we initially conducted Levene’s Test for the t-tests. The obtained p-values for the Levene’s Test were greater than 0.05, suggesting that the variances were not significantly different from one another. Relationships were explored using Spearman correlation analysis. Survival curves were generated using the Kaplan-Meier method and analyzed using Log rank tests. P<0.05 was considered to indicate a statistically significant difference.
Results
SLC31A1 Expression and Intracellular Copper Overload in HGG
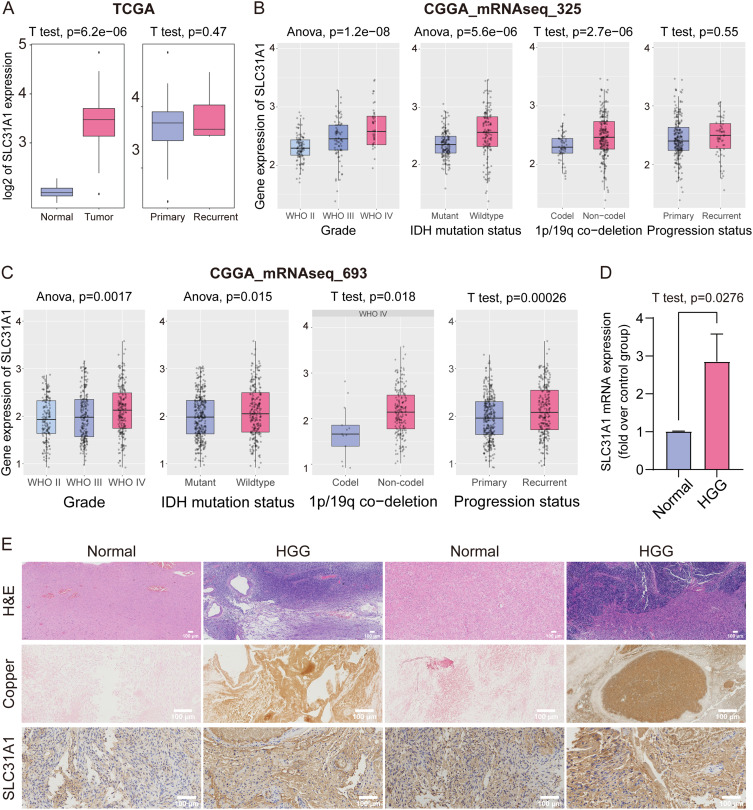
Copper, an indispensable micronutrient crucial for cell growth and survival, has been established as a promoter of tumorigenesis,33 tumor metastasis,8 and PD-L1 expression.7 Previous studies have demonstrated increased intratumoral copper distribution in various cancer types, particularly in brain tumors that exhibit a pronounced affinity for copper.34 To delineate the features of copper homeostasis in glioma, the mRNA expression levels of SLC31A1 in TCGA and CGGA glioma datasets were assessed. The results revealed that SLC31A1 was up-regulated in tumors, correlating with glioma stage, IDH mutation, and 1p/19q co-deletion status (Figure 1A–C). Subsequently, SLC31A1 mRNA expression was assessed in normal and HGG tissues; its expression was upregulated in tumor tissues compared to normal samples (Figure 1D). Similarly, intratumoral copper accumulation was significantly higher in HGG tissues than in normal brain samples, and SLC31A1 exhibited upregulation at the protein level in HGG tissues (Figure 1E). These findings collectively characterize HGG as a malignancy associated with increased copper intake. In recent study published studies,17,18 a distinctive pattern of RCD induced by intracellular copper overload was defined as cuproptosis, identifying 14 key cuproptosis regulated genes (Table S1). Consequently, the present study aimed to explore the impact of cuproptosis on tumor molecular subtyping and analyze the correlation between cuproptosis and therapeutic benefits in HGG, as illustrated in the flow chart (Figure S1).
Figure 1.
Intracellular copper overload and SLC31A1 up-regulation in HGG. (A–C) The expression profile of SLC31A1 in glioma in TCGA and CGGA datasets. (D) qPCR analysis of SLC31A1 mRNA expression in normal tissues (n=3) and HGG tissues (n=4). Data were compared using a Student’s t-test. (E) Histopathological examination using H&E staining, copper salt staining, and IHC staining of HGG and paired normal tissues.
Abbreviations: HGG, high-grade glioma; TCGA, The Cancer Genome Atlas; CGGA, China Glioma Genome Atlas; qPCR, quantitative PCR; H&E, hematoxylin and eosin; IHC, immunohistochemistry.
Genetic Variations and Expression Profiles of CRGs in HGG
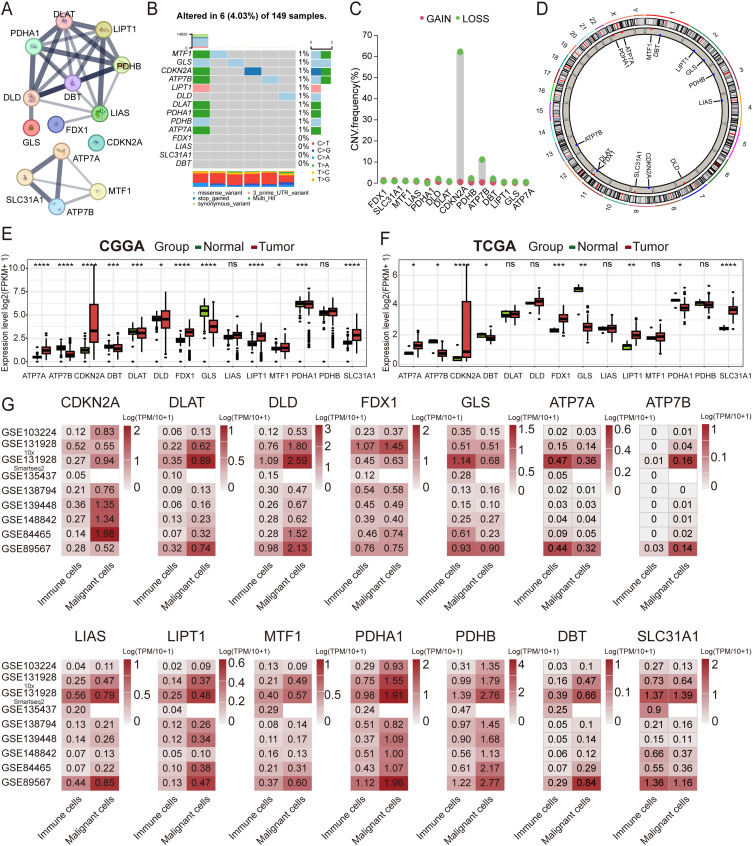
The chromosomal location and coding sequence length of the 14 CRGs were obtained from NCBI (Table S1). The protein-protein interaction network highlighted DBT as the hub gene (Figure 2A). Somatic mutations of the 14 genes revealed that six (4.03%) of the 149 samples exhibited genetic variations (Figure 2B). Copy number variation (CNV) analysis showed a significantly high frequency of copy number deletion (62.26%) for CDKN2A (Figure 2C), with the chromosomal location of CNV alterations visualized in the circos plot (Figure 2D). Differential analysis demonstrated significant upregulation of ATP7A, CDKN2A, FDX1, LIPT1, MTF1, PDHA1 and SLC31A1 in HGG in the CGGA dataset, while ATP7B, DBT, GLS, DLAT, and DLD were down-regulated (Figure 2E). In TCGA, ATP7A, CDKN2A, FDX1, LIPT1 and SLC31A1 were identified as up-regulated genes, whereas ATP7B, DBT, GLS and PDHA1 were significantly down-regulated (Figure 2F). At the protein level, IHC staining downloaded from HPA revealed strong staining for ATP7A, DLD, CDKN2A, and PDHB in HGG (Figure S2A). In primary glioma patients from CGGA, upregulated expression of FDX1 and DLAT was significantly associated with worse OS (Figure S2B and C), while patients with upregulated expression of CDKN2A and GLS exhibited better OS (Figure S2D and E).
Figure 2.
Characterization of the genetic variations and the expression profiles of the 14 CRGs in HGG. (A) Protein-protein interaction network of the 14 CRGs obtained from STRING 11.5 and plotted using the “igraph” R package. (B–D) Somatic mutation signature, CNV frequency, and chromosomal location of the CNVs of the 14 CRGs in the HGG cohort obtained from TCGA. (E and F) Expression profiles of the 14 CRGs in the CGGA and TCGA cohorts. Data were compared using a Student’s t-test, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. (G) Expression of the 14 CRGs at the single-cell level across eight single-cell RNA sequencing datasets downloaded from the TISCH portal.
Abbreviations: HGG, high-grade glioma; CRGs, cuproptosis-related genes; CNV, copy number variation; TCGA, The Cancer Genome Atlas; TISCH, Tumor Immune Single-cell Hub.
At the single-cell level, GLS, ATP7A and SLC31A1 exhibited preferential expression in immune cells, while the other eleven genes were predominantly expressed in malignant cells across eight scRNA-seq datasets (Figure 2G). At the subcellular level in the U251 cell line, ICC staining from HPA suggested that FDX1, DLD, DLAT, PDHB, and GLS were primarily localized in the mitochondria, whereas CDKN2A and MTF1 was predominantly localized in the nucleoplasm (Figure S2F). The cellular and subcellular location and expression patterns of the 14 CRGs in tumor cells suggest that cuproptosis may occur in HGG tumor cells by targeting lipoylated TCA cycle proteins, a phenomenon not previously reported.
Cuproptosis-Related Subtype Identification and Functional Enrichment Analysis
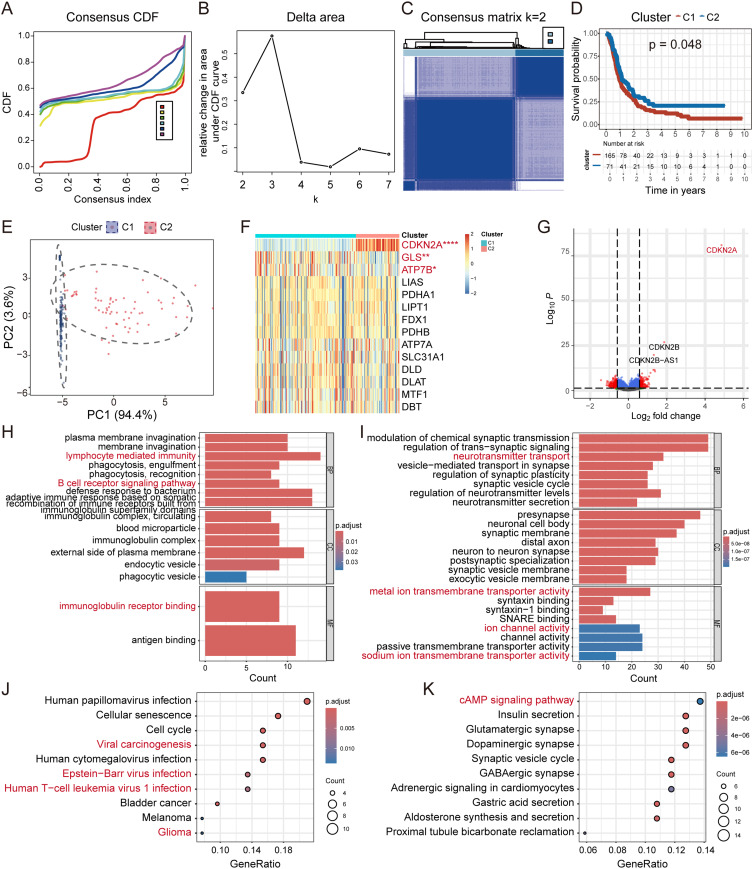
To investigate the association between CRG signatures and HGG subtyping, consensus clustering analysis on the CGGA dataset was employed. Patients were stratified into two groups (K=2) with distinct intergroup correlations (Figure 3A–C). Kaplan-Meier analysis revealed that patients in Cluster 1 (C1) exhibited a worse survival probability (Figure 3D). PCA demonstrated clear differentiation between the two cuproptosis subtypes based on the expression of the 14 CRGs (Figure 3E). No significant differences were observed in IDH mutation status, endpoint event, 1p19q co-deletion status, TMZ status, radio status, or MGMTp methylation status between the groups (Figure S3). In comparison to C1, C2 exhibited a low cuproptosis phenotype with elevated expression of negative hits (CDKN2A and GLS) (Figure 3F).
Figure 3.
Unsupervised clustering and functional enrichment analysis in distinct subgroups of HGG using data obtained from CGGA. (A–C) The “ConsensusClusterPlus” R package was employed to generate the consensus CDF, δ area, and consensus matrix (K=2) in HGG patients using data obtained from CGGA. (D) Kaplan-Meier survival analysis of the two clusters; data were compared using a Log-rank and Wilcoxon-Breslow test. (E) PCA distinguishes the distinct clusters based on the expression of the 14 CRGs. (F) Expression levels of the 14 CRGs between distinct clusters. (G) Volcano plot showing the DEGs between the different clusters (fold change = 1.5). (H–K) GO and KEGG enrichment analysis between the two distinct subgroups of HGG.
Abbreviations: HGG, high-grade glioma; CGGA, China Glioma Genome Atlas; CDF, cumulative distribution function; PCA, principal component analysis; CRG, cuproptosis-related gene; DEG, differentially expressed gene; Go, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
For a more in-depth analysis of these distinct groups, the DEGs with a fold change of |≥1.5| were selected, as shown in the volcano plot (C2 vs C1; Figure 3G). GO analysis of the up-regulated DEGs indicated significant enrichment in biological processes related to lymphocyte mediated immunity (Figure 3H). Down-regulated DEGs were preferentially enriched in activities associated with metal ion transmembrane transport and ion channel regulation (Figure 3I). These findings suggested that C2 exhibited lower ion transport activity, increased phagocytosis, and enhanced humoral immunity. Furthermore, KEGG analysis revealed enrichment in Epstein-Barr virus (EBV) infection and viral carcinogenesis pathways in C2, potentially due to tumor cells in this subgroup being less susceptible to cuproptosis, making them ideal hosts for viruses (Figure 3J). Additionally, GSVA indicated a higher enrichment score for hallmark interferon α and γ response pathways in C2, closely linked to viral infection (Figure S4). A previous study confirmed that increased intracellular copper levels in the host reduced the severity of influenza infection.35 Meanwhile, down-regulated DEGs were significantly enriched in the cAMP signaling pathway and proximal tubule bicarbonate reclamation (Figure 3K). Activation of the cAMP signaling pathway ultimately contributes to HGG cell migration and invasion,36 which may explain the better prognosis observed in C2. To confirm the robustness of the CRGs-based classification, the glioma cohort obtained from TCGA was used for consensus clustering analysis, identifying two subgroups with different OS that were consistent with CGGA (Figure S5A–D).
Characterization of the Immune Landscape in the Cuproptosis-Related Subgroups
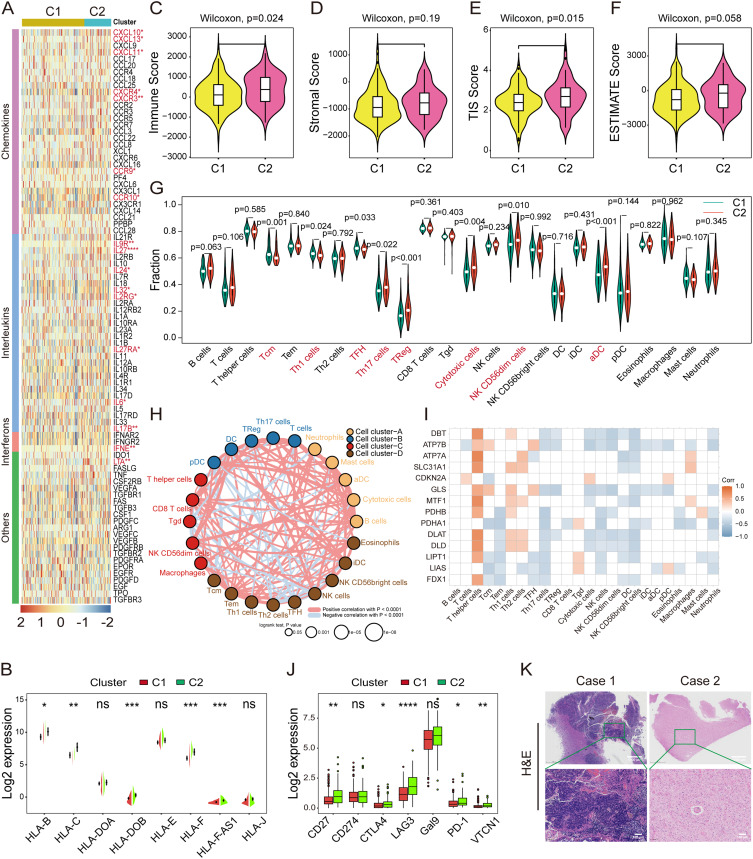
The interplay between cuproptosis and the reprogramming of the TME was investigated next. The heatmap revealed up-regulated expression of chemokines (CXCL10, CXCL11, CXCL13, CXCR2, CXCR3, CXCR4, CXCR9, and CCR10) in C2 compared to C1 (Figure 4A), leading to the recruitment of CD8+ T cells and dendritic cells (DCs). Similarly, interleukins (IL6, IL24, IL27, IL32, IL2RG, IL9R, IL17B, and IL27RA), interferons (IFNE, and LTA), other cytokines (CSF1) (Figure 4A), and the major histocompatibility complexes (MHC; HLA-B, HLA-C, HLA-DOB, HLA-F, and HLA-F-AS1) exhibited higher expression in C2 (Figure 4B). Subsequently, the stromal, immune, ESTIMATE, and TIS scores were compared between C1 and C2, with C2 displaying elevated immune and TIS scores (Figure 4C–F).
Figure 4.
Correlation analysis of cuproptosis and the tumor microenvironment landscape in HGG. (A) Heatmap illustrating the mRNA expression levels of chemokines, interleukins, interferons, and other cytokines in clusters C1 and C2. (B) Split violin plot showing the variations in the MHCs between clusters C1 and C2. Data were compared using a Wilcoxon test. (C–F) Comparison of the stromal, immune, ESTIMATE, and TIS scores between the two clusters. Data were compared using a Wilcoxon test. (G) Infiltration analysis of the 24 immune cell populations in C1 and C2 by ssGSEA. Data were compared using a Wilcoxon test. (H) The immune cell interaction network was assessed using Spearman correlation analysis based on the expression of the 14 CRGs. The line connecting two cells indicates an interaction between them. (I) Correlation analysis between the expression of the 14 CRGs and infiltration of the 24 immune cell populations. (J) mRNA expression levels of immune checkpoints between the two subtypes. Data were compared using a Wilcoxon test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. (K) Representative images of H&E staining depicting two immune phenotypes of HGG.
Abbreviations: HGG, high-grade glioma; MHC, major histocompatibility complex; TIS, Tumor Inflammation Signature; CRG, cuproptosis-related gene; H&E, hematoxylin and eosin.
Among the 24 immune cell populations, Th17 cells, Tregs, cytotoxic cells, NK CD56dim cells, and ɑDC sparsely infiltrated C1, while Tcm and Th1 cells were more abundant in C1 according to ssGSEA (Figure 4G). Furthermore, the immune cell network depicted a comprehensive signature of tumor-immune cell interactions and cell lineages in HGG (Figure 4H). Interestingly, the expression of CRGs was positively correlated with the infiltration of T helper cells and showed a negative association with NK cell infiltration (Figure 4I). Notably, CD27, CTLA-4, LAG3, PD-1, and VTCN1 were significantly down-regulated in C1 (Figure 4J), potentially stemming from the scarce immune cell infiltration in C1. Indeed, these observations revealed two distinct immune phenotypes of HGG through H&E staining, characterized by either abundant or poor immune cell infiltration in tumors (Figure 4K). Together, cuproptosis transformed TME into an immune-desert signature with sparse immune infiltration, lower immune checkpoint expression, and a poorer OS.
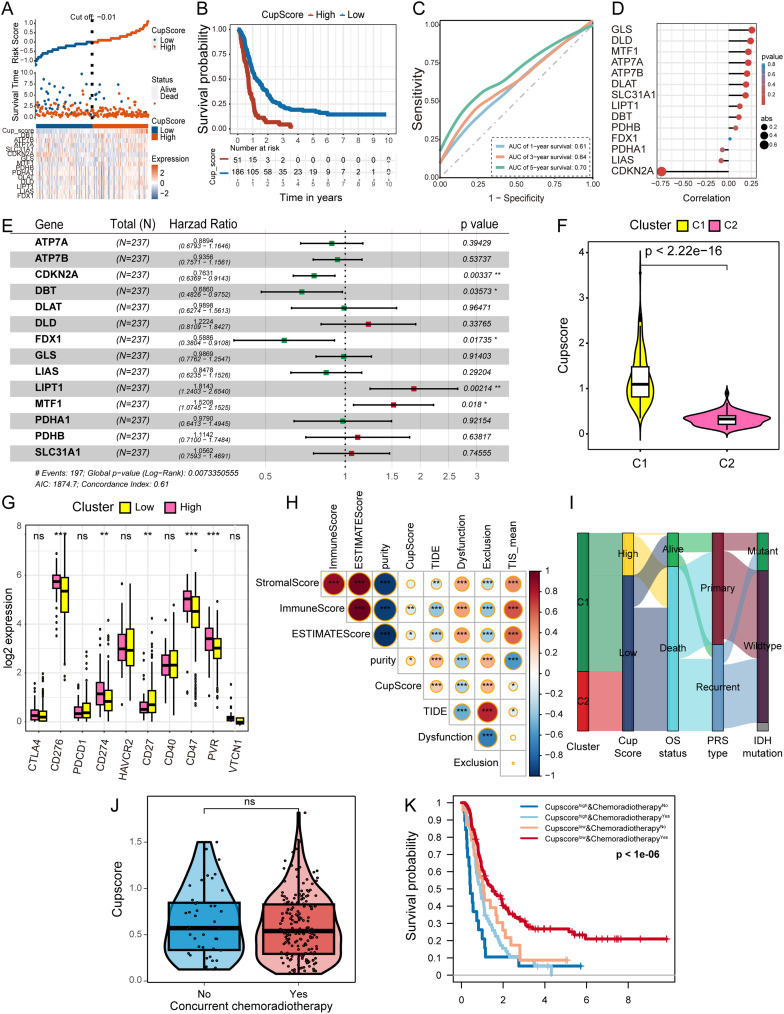
Construction of the CupScore Scoring Model
The above results highlight the pivotal role of cuproptosis in TME remodeling and survival outcomes in HGG. Next, a scoring model named CupScore was developed, derived from the expression patterns of the 14 CRGs. Employing the formula described earlier, patients with HGG were classified into high and low CupScore groups using a cutoff of −0.01 (Figure 5A). Kaplan-Meier analysis showed that patients with higher CupScore experienced worse OS probability (Figure 5B), with an AUC value of 0.70 for the 5-year survival ROC curve (Figure 5C).
Figure 5.
Construction of the CupScore model using the CGGA HGG cohort. (A) Identification of high and low CupScore groups using the “ggrisk” R package with a cutoff value of −0.01. (B) Kaplan-Meier curve illustrating a significant difference in the survival rates between the high and low CupScore groups. (C) ROC curve displaying the AUC values of the CupScore model in predicting the 1, 3, and 5-year survival rates for patients with HGG. (D) Correlation between the CupScore and the expression of the 14 CRGs determined using Spearman correlation analysis. (E) Forest plot illustrating the contributions of the 14 CRGs to the OS of patients with HGG. (F) Violin plot displaying the CupScore distribution in the two clusters. Data were compared using a Wilcoxon test. (G) Expression levels of immune checkpoints between the different CupScore groups. Data were compared using a Student’s t-test. *P<0.05, **P<0.01, ***P<0.001. (H) Heatmap demonstrating the correlation between the CupScore and the ESTIMATE, stromal, TIS, and immune scores. (I) Sankey diagram illustrating the correlation between clusters, CupScore, OS status, PRS type, and IDH mutation status using the “ggalluvial” R package. (J) CupScore between patients who received the standard of care per the Stupp protocol and those who did not. Data were compared using a Wilcoxon test. (K) Kaplan-Meier curve showing a significant difference in the survival between the high and low CupScore groups, both with and without concurrent chemoradiotherapy.
Abbreviations: ROC, receiver operating characteristic; AUC, area under the curve; CGGA, China Glioma Genome Atlas; HGG, high-grade glioma; CRG, cuproptosis-related gene; OS, overall survival.
Exploring the correlation between the CupScore and various clinical features of patients with HGG revealed differences across CRGs-classified clusters (C1 or C2), OS duration (>1 or ≤1 year), and censor status (alive or deceased) (Table S2). Spearman correlation analysis during the model construction process revealed a negative correlation between CDKN2A expression and the CupScore, while GLS and DLD exhibited a positive correlation with the CupScore (Figure 5D). The forest plot highlighted FDX1 as a protective factor for patient OS, while LIPT1 emerged as a high-risk factor (Figure 5E). Notably, patients in C1 demonstrated a significantly higher CupScore compared to those in C2 (Figure 5F). Moreover, a higher CupScore was associated with lower expression of immune checkpoints, including CD276, CD274, CD47, and PVR (Figure 5G). Further analyses indicated that the CupScore was positively correlated with TIDE, tumor purity and exclusion score, whereas it exhibited a negative association with immune, ESTIMATE, dysfunction, and TIS scores (Figure 5H). These findings suggested that cuproptosis actively contributed to TME remodeling, promoting tumor immune tolerance and, was thus consequently associated with a poorer OS in patients with HGG. The Sankey diagram provides a visual representation showing that the majority of patients with a higher a CupScore ultimately succumbed to the tumor (Figure 5I). We compared the CupScore between patients who received the standard Stupp protocol care and those who did not, and found no difference (Figure 5J). Patients who received concurrent chemoradiotherapy with low CupScore showed significantly better OS (Figure 5K), suggesting that CupScore may predict a better response to the standard of care per the Stupp protocol.
Drug Sensitivity Screening in Distinct Subtypes for Chemotherapy Guidance
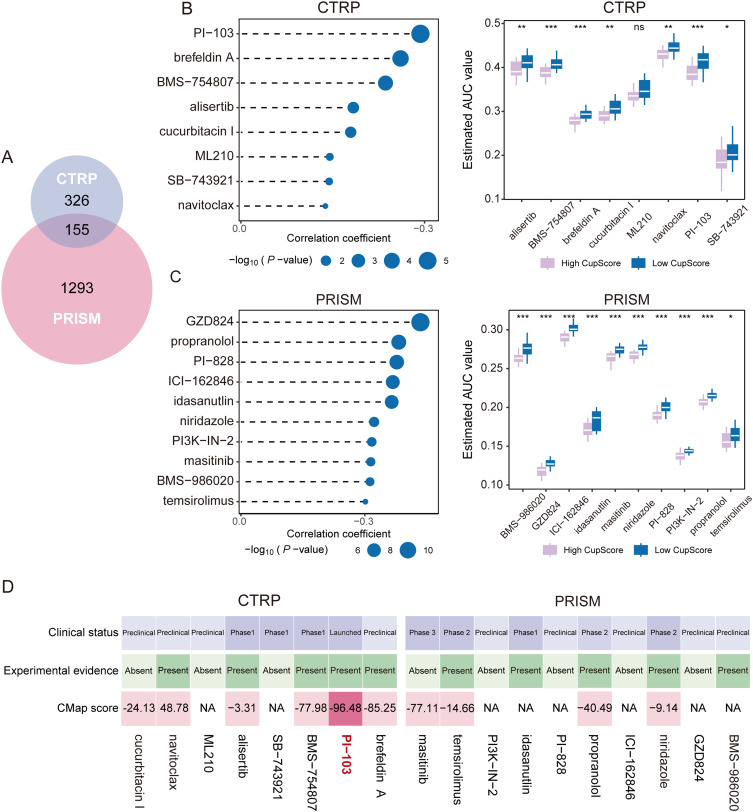
Next, potential therapeutic agents tailored to CRGs-classified clusters were investigated. Initially, the CGGA prediction model on GDSC HGG cancer cells was employed (Figure S6A), comparing the AUC of the drug responses between clusters. Notably, the AUCs of temsirolimus, brivanib, alisertib, and ACY-1215 were significantly lower in C1 (Figure S6B–E).
Subsequently, the CTRP2.0 and PRISM databases were leveraged to identify therapeutic agents for patients with HGG with a high CupScore. Removing duplicates, a total of 1,774 chemical agents were considered (Figure 6A). Spearman correlation analysis between the CupScore and AUC values identified candidate drugs with negative correlation coefficients using the CTRP2.0 and PRISM datasets (Spearman’s r <-0.10 for CTRP or −0.30 for PRISM). This approach led to the identification of eight CTRP-derived drugs (brefeldin A, PI-103, BMS-754807, alisertib, SB-743921, ML210, cucurbitacin I, and navitoclax) and ten PRISM-derived drugs (GZD824, propranolol, PI-828, ICI-162846, idasanutlin, niridazole, PI3K-IN-2, masitinib, BMS-986020, temsirolimus) as candidate agents for patients with HGG with a higher CupScore, exhibiting lower estimated AUC values (Figure 6B and C).
Figure 6.
Identification of candidate drugs for patients with HGG with a high CupScore. (A) Overview of the compounds contained in the CTRP2.0 and PRISM databases illustrated as a Venn diagram. (B and C) Spearman correlation analysis and comparison of drug sensitivity for eight CTRP-derived and nine PRISM-derived drugs. *P<0.05, **P<0.01, ***P<0.001. (D) Integration of experimental evidence, clinical status, expression of drug targets, and CMap scores for CTRP- and PRISM-derived drugs to identify the most promising candidates for patients with high CupScore.
Abbreviations: CMap, Connectivity Map; HGG, high-grade glioma; CTRP, Cancer Therapeutics Response Portal.
To refine the selection among the 18 agents, an in-depth analysis following a methodology outlined in a previous study was performed.37 Initially, the 300 DEGs with the most significant fold changes (150 up-regulated and 150 down-regulated genes) were submitted to the Connectivity Map (CMap) database (https://clue.io/query). This analysis identified PI-103 with a CMap score <-95, signifying its significant efficacy in treating HGG (Figure 6D). Additionally, the fold changes of PI-103 target genes (MTOR, PIK3CA, PIK3CB, PIK3CD, PIK3CG, and PRKDC) in tumors compared to normal sites (Table S3). Finally, by searching the published literature on NCBI, experimental evidence and the clinical trial status of PI-103 for the treatment of HGG were found, confirming its potential for increasing GBM cell apoptosis by inhibiting DNA repair.38 This comprehensive analysis highlighted PI-103 as the most promising therapeutic option for patients with HGG with a higher CupScore.
Role and Value of the CupScore in Predicting ICIs Therapy Benefits
In recent years, tumor immunotherapy has emerged as a promising strategy for eradicating malignant cancers. However, patients with the same cancer types still exhibit different response rates to immunotherapy, potentially due to distinct immune cell infiltration statuses. ICIs therapy is a recognized approach to circumventing immune tolerance and improving immunotherapy benefits for patients.39
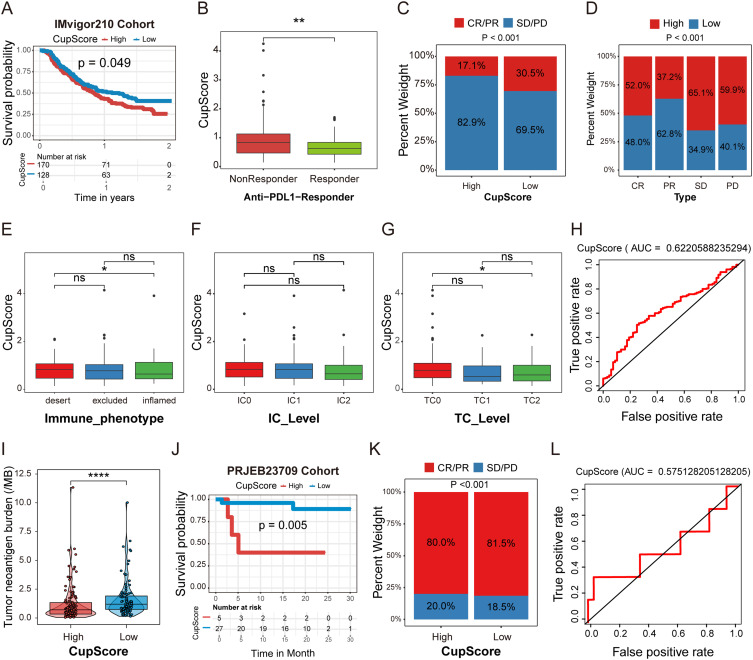
To further explore the relationship between the CupScore and ICIs therapy benefits, an anti-PD-L1 therapy cohort (IMvigor210) of urothelial carcinoma was investigated. A total of 298 patients undergoing anti-PD-L1 therapy were clustered into high (n=170) and low (n=128) CupScore groups, with patients in the low CupScore group exhibiting a higher survival probability rate (Figure 7A). Notably, patients in the low CupScore group were more likely to respond to anti-PD-L1 therapy (Figure 7B) and achieved a higher complete response (CR)/partial response (PR) rate (Figure 7C and D).
Figure 7.
CupScore for predicting the responsiveness of patients with HGG to ICIs therapy. (A) Kaplan-Meier survival analysis comparing the different CupScore groups in the IMvigor210 cohort. (B) Comparison of the CupScore between the anti-PD-L1 responder and non-responder groups. Data were compared using a Wilcoxon test. **P<0.01. (C and D) CR/ PR and SD/PD rates between the different CupScore groups following anti-PD-L1 therapy. (E–G) Correlation between the CupScore and immune phenotype, IC, and TC levels. Tumor samples were scored using IHC based on PD-L1 expression in TCs and ICs. *P<0.05. (H) ROC curve evaluating the accuracy of the CupScore in predicting anti-PD-L1 ICI therapy response. (I) TMB comparison between high and low CupScore groups. Data were compared using a Wilcoxon test. ****P<0.0001. (J) Kaplan-Meier survival analysis between the high and low CupScore groups in the PRJEB23709 cohort. (K) CR/PR and SD/PD rates between the different CupScore groups following anti-PD-1 and anti-CTLA4 combination therapy. (L) ROC curve assessing the accuracy of the CupScore in predicting anti-PD-1 and anti-CTLA4 combination ICIs therapy response.
Abbreviations: HGG, high-grade glioma; ICIs, immune checkpoint inhibitors; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; IC, immune cell; TC, tumor cell; IHC, immunohistochemistry; ROC, receiver operating characteristic curve; TMB, tumor mutation burden.
Across most solid tumors, three distinct immunological phenotypes prevail: Immune-desert, immune-excluded, or immune-inflamed, characterized by the infiltration numbers and location of CD8+ T cells in tumors.40 In the IMvigor210 cohort, the relationship between the CupScore and immune phenotype, tumor-infiltrating immune cells (IC) level, and tumor cells (TC) level was explored. The box plot revealed that patients with a higher CupScore displayed an immune-desert phenotype (Figure 7E). Furthermore, specimens scored as IHC TC0 exhibited a higher CupScore than the other two groups (Figure 7F and 7G), strongly suggesting that the CupScore can predict the response to anti-PD-L1 therapy. The ROC curve indicated that the AUC value of the CupScore model for predicting the anti-PD-L1 response was 0.62 (Figure 7H).
Interestingly, a higher CupScore was closely associated with a lower tumor mutation burden (TMB) (Figure 7I). Previous studies have consistently demonstrated that a higher TMB results in an improved response to ICIs therapy across various types of cancer,41 suggesting that the CupScore may serve as a valuable biomarker for assessing the effectiveness of ICIs therapy. Moreover, our model was applied to the PRJEB23709 cohort to explore the correlation between the CupScore and anti-PD-1 and CTLA-4 combination immunotherapy benefits. Patients with melanoma following anti-PD-1 and CTLA-4 combination therapy were divided into high (n=5) and low (n=27) CupScore groups; patients with a higher CupScore exhibited worse OS rates (Figure 7J). Similarly, patients with a lower CupScore showed a higher CR/PR rate in the PRJEB23709 cohort (Figure 7K), and the AUC value of the model was 0.45 (Figure 7L). Together, the above results indicate that cuproptosis reprograms the TME, influencing the immune phenotype of HGG. The CupScore model may thus serve as a powerful metric for predicting the response to ICIs therapy.
CDKN2A facilitates the proliferation and migration of HGG
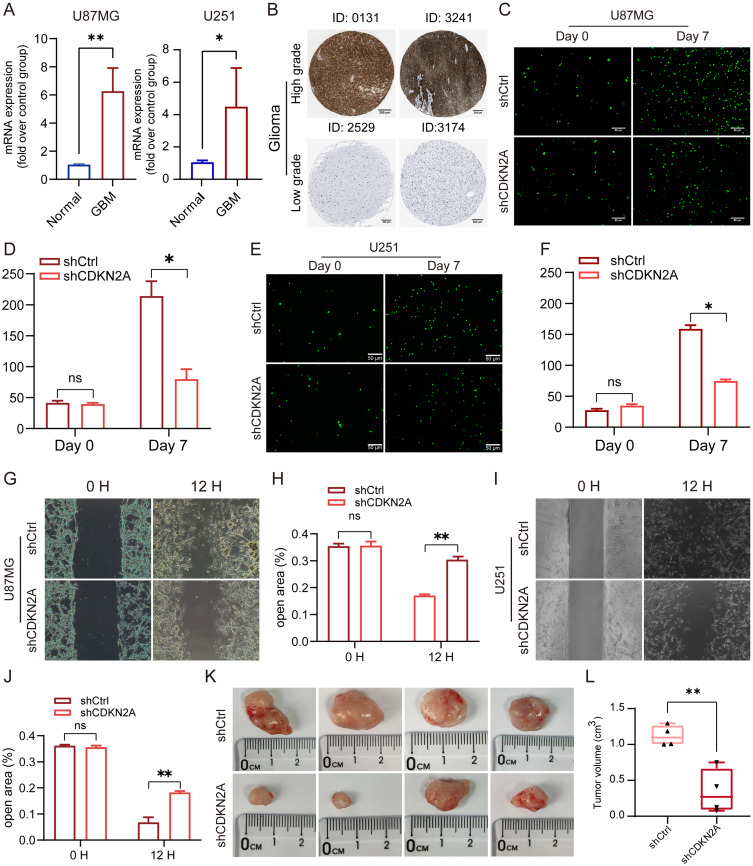
Significantly higher expression of CDKN2A was observed at both the mRNA level (Figure 2E and F) and protein level (Figure S2A) in HGG. Consequently, in vivo and in vitro experiments were performed to explore the potential biological function of CDKN2A in HGG. CDKN2A was confirmed to be upregulated in U87MG and U251 cells at the mRNA level (Figure 8A) and exhibited higher expression in higher-grade glioma (Figure 8B). The cell proliferation assay indicated that CDKN2A knockdown using shCDKN2A reduced the proliferation and migration of the U87MG and U251 cell lines (Figure 8C–J). Additionally, U87MG cells with CDKN2A expression knocked down exhibited smaller tumor volumes in the mouse model (Figure 8K and L). In conclusion, CDKN2A plays a significant role in HGG′ malignant phenotype development.
Figure 8.
CDKN2A promotes the proliferation and migration of HGG cells. (A) Upregulated expression of CDKN2A in U87MG and U251 cell lines were detected by qPCR. (B) CDKN2A expression is upregulated in high-grade glioma compared to low-grade glioma as observed using IHC images from HPA. (C–J) U87MG and U251 cells transfected with shCDKN2A exhibited reduced proliferation and migration compared with cells transfected with the shCtrl. (K–L) Comparison of the tumor size between the control tumors and tumors with CDKN2A knockdown. All experiments were repeated at least three times, and data were compared using a Student’s t-test. *P<0.05, **P<0.01.
Abbreviations: HGG, high-grade glioma; qPCR, quantitative PCR; IHC, Immunohistochemistry; HPA, Human Protein Atlas; sh, short hairpin.
Discussion
HGG is characterized by a poor prognosis and high intertumoral and intratumoral heterogeneity, with a lack of effective therapies. To date, various molecular classification strategies have been proposed to develop innovative targeted therapies for individual subtypes, such as those based on transcriptomic profiles, DNA methylation, and somatic genomic alterations.42 However, existing HGG subtyping strategies have not yielded apparent benefits for patients, highlighting the urgent need for more effective molecular subtyping methods to tailor therapies and improve patient outcomes.
Copper is an essential micronutrient for most organisms, and imbalances in copper homeostasis are commonly observed in the serum or tissue of patients with cancer.43 Excess intracellular copper induces a recently defined form of RCD known as cuproptosis, which is caused by copper-induced lipoylated protein aggregation and loss of iron-sulfur cluster proteins.17 Copper overloading is the primary requirement and trigger for cuproptosis in tumor cells; however, no concrete evidence has implicated the definitive role of cuproptosis in HGG molecular subtyping, TME reprogramming, or the prediction of therapeutic benefits. Tumors, especially brain tumors, exhibit a higher demand for copper to support cell growth and proliferation. In the present study, upregulated expression of SLC31A1 and excess copper accumulation in HGG was observed using qPCR, IHC staining, and copper salt staining. Due to their higher copper-dependent characteristics, brain tumor cells are more vulnerable to copper overload and copper-induced toxicity, highlighting copper regulatory mechanisms as potential therapeutic targets for HGG treatment. The present study comprehensively investigated the role of CRGs in HGG subtyping and TME reprogramming. Additionally, a CRG-based scoring model, termed CupScore, was constructed to explore the association between the CupScore with chemotherapy and ICI therapy benefits in patients with HGG.
In the present study, patients were divided into two clusters (C1 and C2) based on the expression of 14 CRGs in the CGGA training cohort and subsequently validated in the data obtained from TCGA. PCA confirmed that the two distinct cuproptosis-related subtypes could be clearly distinguished by the expression of the 14 CRGs. Patients in C1 with a higher CupScore exhibited worse survival rates compared to those in C2. In the GO and KEGG analysis between the two groups, C2 showed high enrichment in phagocytosis, EBV infection, and viral carcinogenesis pathways, possibly resulting from lower sensitivity to cuproptosis. Meanwhile, patients in C2 exhibited suppressed metal ion transmembrane transporter activity and ion channel regulator activity, suggesting less copper accumulation in C2 tumors. A previous study documented that host copper overloading decreased influenza infection,35 which may explain why tumors in C2 were more susceptible to EBV infection.44 In summary, tumors in C2 with lower sensitivity to cuproptosis were more likely to be associated with viral infection.
The present study illustrated a significant association between cuproptosis levels and TME remodeling in HGG. Firstly, tumors in C2 had higher immune scores with an abundance of cytotoxic cells, Th17 cells, NK CD56dim cells, and ɑDCs. Numerous studies have reported that the abundance of immune cells in tumor sites affected antitumor immunity and immunotherapy efficacy.45 Further, it was found that patients in C2 exhibited higher TIS scores, as well as higher levels of chemokines, interleukins, interferons, cytokines, and the MHC complex, which contributed to the recruitment of CD8+ T cells, DCs, NK cells, and NKT cells to kill tumor cells. It has been previously documented that patients with higher TIS scores exhibit prolonged OS compared to patients with lower scores and that the TIS score was an independent and accurate predictive biomarker for the evaluation of the clinical benefit of ICIs therapy.46 Of note, tumors in C2 expressed high levels of immune checkpoint proteins (CD27, CTLA-4, LAG3, PD-1, and VTCN1), which are associated with the response to ICIs therapy.
Considering the crucial role of cuproptosis in TME remodeling and the heterogeneity amongst the CRG-related subgroups of HGG, a scoring system, CupScore, was developed to evaluate the value of assessing cuproptosis in predicting the response to ICIs therapy, as well as to identify promising candidate drugs for patients with a higher CupScore. In the IMvigor210 cohort, patients with lower cuproptosis scores showed an immune-inflamed phenotype with abundant infiltration of immune cells, high expression of immune checkpoints, and a longer survival time when treated with anti-PD-L1 therapy. In addition, there was a positive correlation between the CupScore and TMB. A higher TMB resulted in a greater variety and quantity of tumor neoantigen burden, increasing the likelihood of T cell recognition and thus there was a clinical correlation with improved ICIs therapy outcomes.47 Furthermore, the CupScore model also showed remarkable value in predicting the benefits of anti-PD-1 and CTLA-4 combination therapy. The relationship between the CupScore and the AUC value of drugs was assessed using Spearman correlation analysis, and PI-103 was identified as the most promising candidate compound for the treatment of tumors with a high CupScore. Experimental evidence uncovered that PI-103 promoted HGG cell line apoptosis by inhibiting DNA repair and is currently being assessed in a clinical trial. In conclusion, the CupScore model is a promising metric for predicting the response to ICIs therapy and chemotherapy, highlighting a novel perspective for the clinical treatment of HGG. Furthermore, patients who received concurrent chemoradiotherapy with a low CupScore showed significantly better overall survival (OS), suggesting that CupScore may predict the response to the standard of care according to the Stupp protocol.
In vivo and in vitro experiments were used to explore the potential biological function of CDKN2A in HGG and it was found that CDKN2A promoted the proliferation and migration of U87MG and U251 cells. Moreover, subcutaneous tumor growth assays showed that CDKN2A contributed to an increased tumor burden in a nude mouse model. Taken together, CDKN2A played an essential role in the formation of a malignant phenotype during the progression of HGG.
Despite the extensive and systematic analysis performed in the present study, there remain some limitations. First, as a relatively recently defined type of RCD, only 14 CRGs were identified for unsupervised tumor clustering. Therefore, more CRGs with higher accuracy are required for improved tumor subtyping and score model construction. Second, considering the impact of the hard-to-reconcile batch effect on subtyping, only one HGG cohort was selected from the CGGA dataset for further analysis. In the future, larger cohorts from several centers will be assessed for further in-depth analysis and verification. Finally, besides constructing signatures and identifying molecular subtypes for HGG based on CRG-based signature construction and molecular subtyping, approaches such as the ferroptosis-related model and multi-omics profiling can also be considered as potential alternatives.48,49
In conclusion, the present study delineated two distinct cuproptosis-related molecular subtypes based on the expression of 14 CRGs in HGG, revealing divergent prognoses. CRGs play a pivotal role in tumor heterogeneity and TME remodeling in the HGG, and their expression is linked to the benefits of chemotherapy and ICIs therapy. The CupScore model may serve as a promising tool capable of characterizing molecular subtype profiles and TME landscapes, predicting responses to chemotherapy and immunotherapy, and assessing the long-term survival rates of patients with HGG.
Funding Statement
The present study was supported by funding from the Scientific Research Project of the Nantong Municipal Health Commission (grant no. MS2022048) and the Huaian Science and Technology Project (grant no. HAB202138).
Data Sharing Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of the Huaian Cancer Hospital (No. 2019036) and conformed to the guidelines described in the Declaration of Helsinki. Informed patient consent was obtained from all patients prior to inclusion.
Consent for Publication
All authors have read and approved the final manuscript.
Author Contributions
Xiaochun Xia, Pengqin Xu and Peng Li took part in drafting, revising or critically reviewing the article; Xiaoying Huang and Longxiang Wu participated in data analysis and experiments validation. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Weller M, Wen PY, Chang SM. et al. Glioma. Nat Rev Dis Primers. 2024;10(1):33. doi: 10.1038/s41572-024-00516-y [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231–1251. doi: 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitfield BT, Huse JT. Classification of adult-type diffuse gliomas: impact of the World Health Organization 2021 update. Brain Pathol. 2022;32(4):e13062. doi: 10.1111/bpa.13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14(1):48. doi: 10.1186/s12943-015-0321-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–296. doi: 10.1038/s41571-020-00462-0 [DOI] [PubMed] [Google Scholar]
- 6.Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol. 2008;4(3):176–185. doi: 10.1038/nchembio.72 [DOI] [PubMed] [Google Scholar]
- 7.Voli F, Valli E, Lerra L, et al. Intratumoral Copper Modulates PD-L1 Expression and Influences Tumor Immune Evasion. Cancer Res. 2020;80(19):4129–4144. doi: 10.1158/0008-5472.CAN-20-0471 [DOI] [PubMed] [Google Scholar]
- 8.Ramchandani D, Berisa M, Tavarez DA, et al. Copper depletion modulates mitochondrial oxidative phosphorylation to impair triple negative breast cancer metastasis. Nat Commun. 2021;12(1):7311. doi: 10.1038/s41467-021-27559-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang D, Chen X, Kroemer G. Cuproptosis: a copper-triggered modality of mitochondrial cell death. Cell Res. 2022. doi: 10.1038/s41422-022-00653-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106(20):8344–8349. doi: 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeCordova S, Shastri A, Tsolaki AG, et al. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front Immunol. 2020;11:1402. doi: 10.3389/fimmu.2020.01402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Lu Z, Wang M, et al. The cuproptosis-related signature associated with the tumor environment and prognosis of patients with glioma. Front Immunol. 2022;13:998236. doi: 10.3389/fimmu.2022.998236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong W, Li C, Wan B, et al. N6-Methyladenosine Regulator-Mediated Immue Patterns and Tumor Microenvironment Infiltration Characterization in Glioblastoma. Front Immunol. 2022;13:819080. doi: 10.3389/fimmu.2022.819080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu W, Ma Y, Hou W, et al. Identification of Immune-Related lncRNA Prognostic Signature and Molecular Subtypes for Glioblastoma. Front Immunol. 2021;12:706936. doi: 10.3389/fimmu.2021.706936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Gao L, Guo X, et al. Development and validation of a nomogram with an autophagy-related gene signature for predicting survival in patients with glioblastoma. Aging. 2019;11(24):12246–12269. doi: 10.18632/aging.102566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao D, Zhou Y, Wang X, Zhao H, Nie C, Jiang X. A Ferroptosis-Related Prognostic Risk Score Model to Predict Clinical Significance and Immunogenic Characteristics in Glioblastoma Multiforme. Oxid Med Cell Longev. 2021;2021(1):9107857. doi: 10.1155/2021/9107857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–1261. doi: 10.1126/science.abf0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen P, Han H, Wang X, Wang B, Wang Z. Novel Cuproptosis-Related Gene Signature for Precise Identification of High-Risk Populations in Low-Grade Gliomas. Mediators Inflamm. 2023;2023:6232620. doi: 10.1155/2023/6232620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang D, Yu Z, He Y, et al. Strain-Insensitive Elastic Surface Electromyographic (sEMG) Electrode for Efficient Recognition of Exercise Intensities. Micromachines. 2020;11(3):239. doi: 10.3390/mi11030239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abd ElHafeez S, Torino C, D’Arrigo G, et al. An overview on standard statistical methods for assessing exposure-outcome link in survival analysis (Part II): the Kaplan-Meier analysis and the Cox regression method. Aging Clin Exp Res. 2012;24(3):203–206. doi: 10.1007/BF03325249 [DOI] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu S, Hu E, Cai Y, et al. Using clusterProfiler to characterize multiomics data. Nat Protoc. 2024;19(11):3292–3320. doi: 10.1038/s41596-024-01020-z [DOI] [PubMed] [Google Scholar]
- 25.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61(1):92–105. doi: 10.1111/j.0006-341X.2005.030814.x [DOI] [PubMed] [Google Scholar]
- 26.Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol. 2017;17(1):53. doi: 10.1186/s12874-017-0332-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith HG, Jamal K, Dayal JH, et al. RIPK1-mediated immunogenic cell death promotes anti-tumour immunity against soft-tissue sarcoma. EMBO Mol Med. 2020;12(6):e10979. doi: 10.15252/emmm.201910979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scire J, Huisman JS, Grosu A, et al. estimateR: an R package to estimate and monitor the effective reproductive number. BMC Bioinf. 2023;24(1):310. doi: 10.1186/s12859-023-05428-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Mao X, Ye L, Cheng H, Dai X. The Role of the S100 Protein Family in Glioma. J Cancer. 2022;13(10):3022–3030. doi: 10.7150/jca.73365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao P, Wang H, Li H, et al. miR-21-5p Inhibits the Proliferation, Migration, and Invasion of Glioma by Targeting S100A10. J Cancer. 2023;14(10):1781–1793. doi: 10.7150/jca.84030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo J, Cheng J, Zheng N, et al. Copper Promotes Tumorigenesis by Activating the PDK1-AKT Oncogenic Pathway in a Copper Transporter 1 Dependent Manner. Adv Sci. 2021;8(18):e2004303. doi: 10.1002/advs.202004303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denoyer D, Masaldan S, La Fontaine S, Cater MA. Targeting copper in cancer therapy: ‘Copper That Cancer’. Metallomics. 2015;7(11):1459–1476. doi: 10.1039/C5MT00149H [DOI] [PubMed] [Google Scholar]
- 35.Puchkova LV, Kiseleva IV, Polishchuk EV, Broggini M, Ilyechova EY. The Crossroads between Host Copper Metabolism and Influenza Infection. Int J Mol Sci. 2021;22(11):5498. doi: 10.3390/ijms22115498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang K, Yao G, Hu L, et al. MOB2 suppresses GBM cell migration and invasion via regulation of FAK/Akt and cAMP/PKA signaling. Cell Death Dis. 2020;11(4):230. doi: 10.1038/s41419-020-2381-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang C, Huang X, Li Y, Chen J, Lv Y, Dai S. Prognosis and personalized treatment prediction in TP53-mutant hepatocellular carcinoma: an in silico strategy towards precision oncology. Brief Bioinform. 2021;22(3):1. [DOI] [PubMed] [Google Scholar]
- 38.Westhoff MA, Kandenwein JA, Karl S, et al. The pyridinylfuranopyrimidine inhibitor, PI-103, chemosensitizes glioblastoma cells for apoptosis by inhibiting DNA repair. Oncogene. 2009;28(40):3586–3596. doi: 10.1038/onc.2009.215 [DOI] [PubMed] [Google Scholar]
- 39.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18(3):197–218. doi: 10.1038/s41573-018-0007-y [DOI] [PubMed] [Google Scholar]
- 40.Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. doi: 10.1093/annonc/mdy495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee E, Yong RL, Paddison P, Zhu J. Comparison of glioblastoma (GBM) molecular classification methods. Semin Cancer Biol. 2018;53:201–211. doi: 10.1016/j.semcancer.2018.07.006 [DOI] [PubMed] [Google Scholar]
- 43.Kuo MT, Fu S, Savaraj N, Chen HH. Role of the human high-affinity copper transporter in copper homeostasis regulation and cisplatin sensitivity in cancer chemotherapy. Cancer Res. 2012;72(18):4616–4621. doi: 10.1158/0008-5472.CAN-12-0888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zavala-Vega S, Castro-Escarpulli G, Hernandez-Santos H, et al. An overview of the infection of CMV, HSV 1/2 and EBV in Mexican patients with glioblastoma multiforme. Pathol Res Pract. 2017;213(3):271–276. doi: 10.1016/j.prp.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 45.Helmink BA, Reddy SM, Gao J, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. doi: 10.1038/s41586-019-1922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Damotte D, Warren S, Arrondeau J, et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J Transl Med. 2019;17(1):357. doi: 10.1186/s12967-019-2100-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell. 2021;39(2):154–173. doi: 10.1016/j.ccell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du Y, Li R, Fu D, et al. Multi-omics technologies and molecular biomarkers in brain tumor-related epilepsy. CNS Neurosci Ther. 2024;30(4):e14717. doi: 10.1111/cns.14717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y, Du Y, Zheng Q, et al. Identification of Ferroptosis-Related Prognostic Signature and Subtypes Related to the Immune Microenvironment for Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Front Immunol. 2022;13:895110. doi: 10.3389/fimmu.2022.895110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.