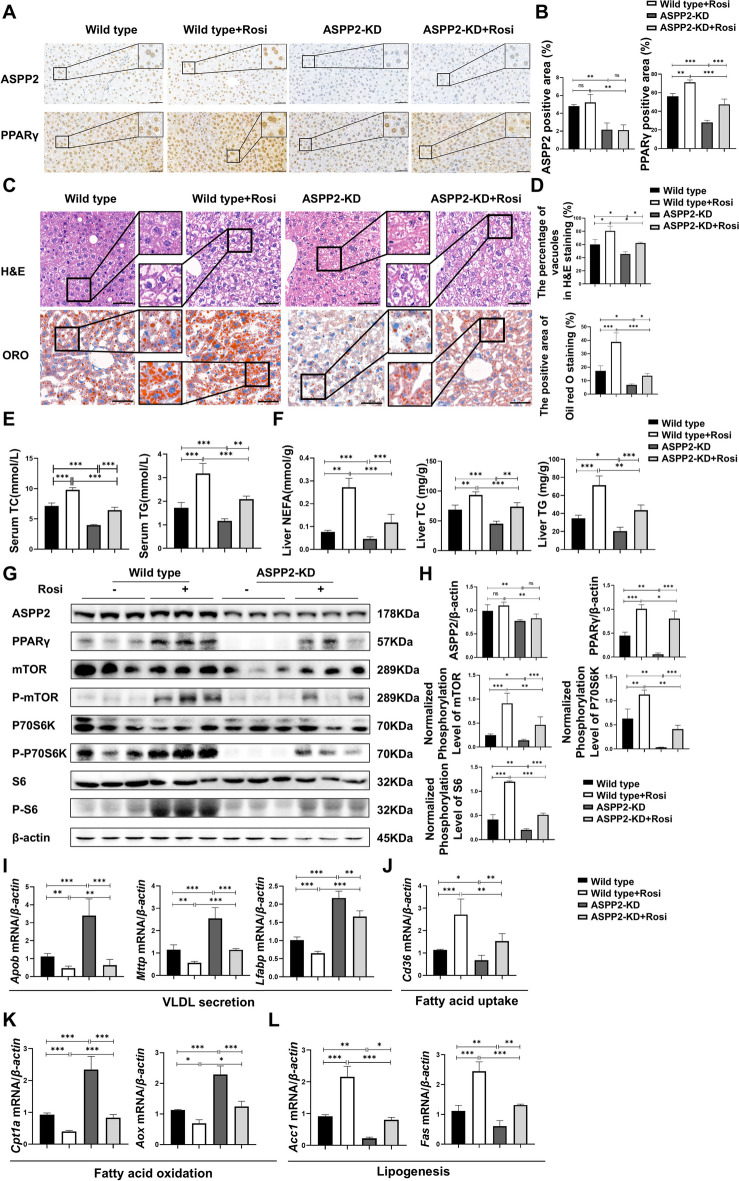
Fig. 4.
ASPP2 inhibits lipid metabolism through the PPARγ-mTOR signaling pathway in alcohol-induced mice models. (A and B) Representative IHC analysis of ASPP2 (top) and PPARγ (bottom) in liver sections, scar bar = 50 µm (A) and quantification analyses (B) of ASPP2 and PPARγ positive area (%) in mouse liver tissue from wild-type mice and ASPP2-KD mice with or without rosiglitazone (Rosi). The extended part of the black lines shows the enlarged image from the black box area. (C) Representative H&E staining (top) and Oil red O staining (bottom) of liver tissue, scar bar = 50 µm. (D) Analyses of the percentage of vacuoles in H&E staining (top) and the positive area in oil red O staining (bottom). (E) Serum TC and TG levels. (F) Liver NEFA, TC and TG levels. (G and H) western blot (G) and quantification analyses (H) of ASPP2, PPARγ, phospho-mTOR, phospho-S6, phospho-p70S6K, and β-actin in mouse liver tissue from wild-type mice and ASPP2-KD mice with or without Rosi. (I, J, K, and L) The mRNA levels of genes involved in lipid metabolism in mouse liver tissue from wild-type mice and ASPP2-KD mice with or without Rosi measured by real-time PCR. The values represent the means ± SEMs (n = 6 in each group). nsP > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. Independent-samples T tests between two groups and one-way ANOVA followed by Bonferroni post hoc test for multiple comparisons were used for statistical analyses