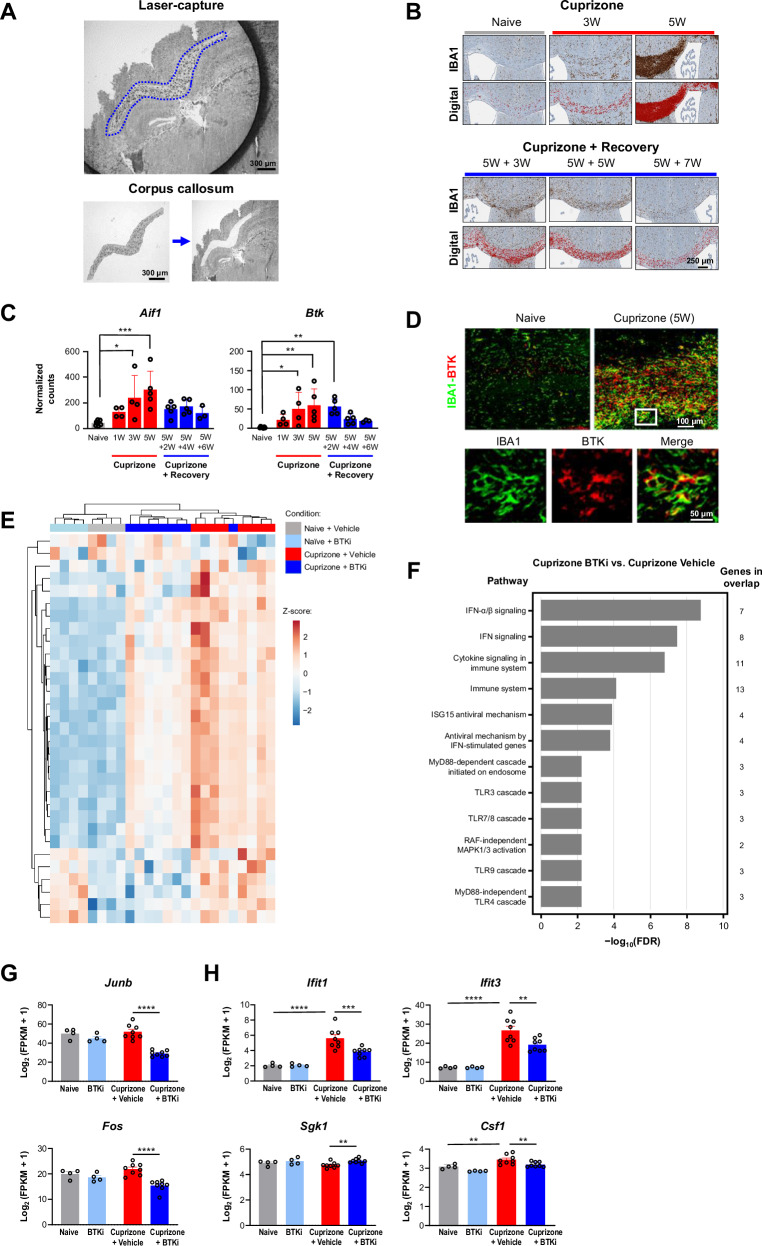
Fig. 2. BTKi in a cuprizone mouse model of demyelination.
A Isolation of corpus callosum from mouse brain using laser-capture microdissection. B Immunostaining for microglial/macrophage marker IBA1 was used to generate a digital signal in HALO. This experiment was reproduced twice with similar results. C DESeq2 normalized counts for the Btk gene and the microglia/macrophage marker gene Aif1 during cuprizone treatment and subsequent recovery (n = 3–7 mice). P-values for Aif1: naive vs. 3W and 5W are 0.012 and 0.0003, respectively. P-values for Btk: naive vs. 3W, 5W, and 5W + 2W are 0.0252, 0.0029, and 0.0053, respectively. D Fluorescent immunostaining for BTK and IBA1. Mouse BTK histology was the result of many experiments to determine optimal antibodies and concentrations to selectively stain for BTK+ cells. E Heatmap of PRN2675-reversed transcriptional signature genes. Heatmap displays log2(FPKM + 1) data scaled using a Z-score. Differential expression analysis was performed in Array Studio using a general linear model. The PRN2675-reversed transcriptional signature consists of 31 differentially expressed genes (absolute(fold-change) ≥ 1.2 and p-value ≤ 0.05) in Cuprizone + Vehicle vs Naive + Vehicle that were reversed in Cuprizone + PRN2675 vs Cuprizone + Vehicle. F Pathway analysis of genes identified in panel E, performed using EnrichR tool based on 2022 Reactome Database. The pathway analysis gene list is the PRN2675-dependent transcriptional signature identified in panel (E). G Expression of the Junb and Fos transcription factor genes quantified as log2(FPKM + 1). Shown p-values were calculated using Array Studio (n = 4–8 mice). P-values are available in the Source Data file. H Expression of the Ifit1,Ifit3, Sgk1, and Csf1 genes quantified as log2(FPKM + 1). Shown p-values were calculated using Array Studio (n = 4–8 mice). P-values are available in the Source Data file. P-values are indicated by * ≤ 0.05, ** ≤ 0.01, *** ≤ 0.001, and **** ≤ 0.0001. Data are presented as mean values ± SEM. BTK Bruton’s tyrosine kinase, BTKi BTK inhibitor, FDR false discovery rate, FPKM fragments per kilobase of transcript per million mapped reads, IBA1 ionized calcium binding adapter molecule 1, IFN interferon, MAPK mitogen-activated protein kinase, RAF rapidly accelerated fibrosarcoma kinase, W week mitogen-activated protein kinase, RAF rapidly accelerated fibrosarcoma kinase, W week. Source data are provided as a Source Data file.