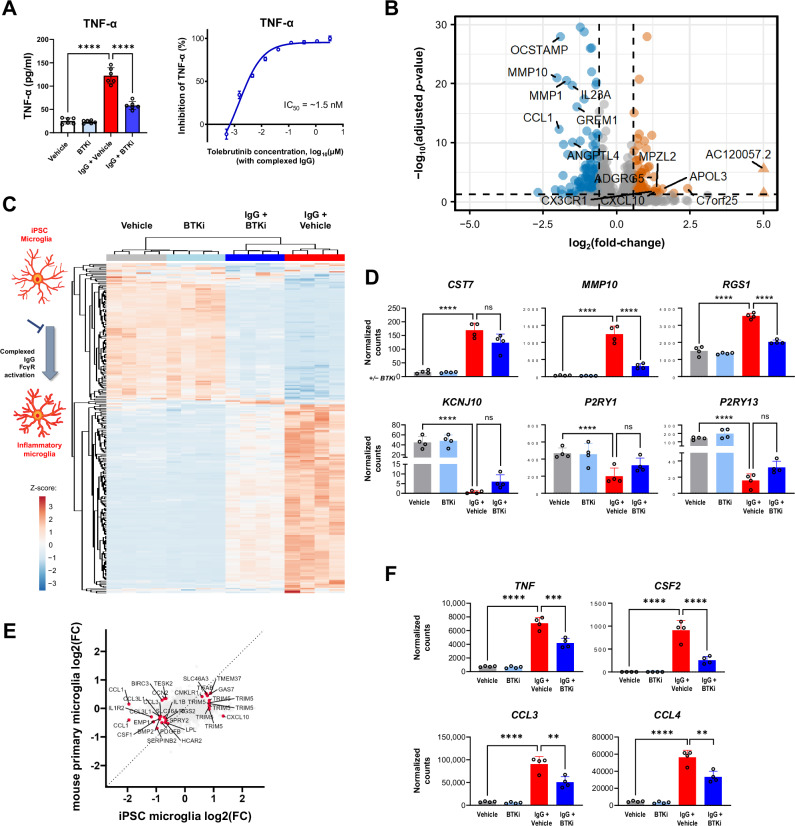
Fig. 4. Identification of a BTK-dependent transcriptional signature in human iPSC-derived microglia.
A Effect of tolebrutinib on TNF-α secretion in human iPSC-derived microglia, 24 h after complexed IgG stimulation (10 μg/mL Fc OxyBURST™; Invitrogen, F2902). Shown p-values were <0.0001 and calculated using one-way ANOVA with post hoc Sidak test (left panel n = 6 technical replicates, right panel n = 3–6 technical replicates). B Volcano plot of differential expression between tolebrutinib-treated and untreated hiPSC-derived microglia, 6 h after complexed IgG stimulation. Differential expression analysis was performed using DESeq2. 250 genes were observed to be differentially expressed in the IgG + tolebrutinib condition (absolute(fold-change) ≥1.5 and FDR ≤ 0.05). The 14 most strongly up- and down-regulated genes (as ranked by log2(fold-change)) are labeled (Wald significance test implemented in DESeq2). C Heatmap of the 250 tolebrutinib-specific transcriptomic signature identified in (B). Heatmap displays DESeq2 normalized counts scaled using a Z-score. D Effect of tolebrutinib 100 nM on expression levels of the CST7, MMP10, and RGS1 genes, which were induced by complexed IgG, and the KCN1J10, P2RY1, and P2RY13 genes, which were inhibited in response to complexed IgG. DESeq2 normalized counts are shown, with FDR-corrected p-values generated using DESeq2 (Wald significance test implemented in DESeq2, n = 4 technical replicates). P-values are available in the Source Data file. E Four-way plot of hiPSC-derived microglia differential expression results for IgG + tolebrutinib vs IgG (Fig. 4B) and primary mouse microglia differential expression results for IgG + tolebrutinib vs IgG (Fig. 3G). Labeled genes were significantly differentially expressed genes in common between the two analyses (for hiPSC-derived microglia, absolute(fold-change) ≥1.5 and FDR ≤ 0.05; for mouse microglia, absolute(fold-change) ≥1.2 and p value ≤ 0.05) (Wald significance test implemented in DESeq2). F DESeq2 normalized counts of genes encoding proinflammatory cytokines and chemokines in hiPSC-derived microglia. Shown p-values are FDR-corrected and were calculated using DESeq2 (Wald significance test implemented in DESeq2, n = 4 technical replicates). P-values are available in the Source Data file. P-values are indicated by * ≤ 0.05, ** ≤ 0.01, *** ≤ 0.001, and **** ≤ 0.0001. Data are presented as mean values ± SEM. BTK Bruton’s tyrosine kinase, IC50 half-maximal inhibitory concentration, hiPSC human induced pluripotent stem cell, IgG immunoglobulin G, mus. mouse, ns non-significant, TNF-α tumor necrosis factor-alpha. Source data are provided as a Source Data file.