Abstract
Background
The intricate interplay between stemness markers and cell death pathways significantly influences the pathophysiology of cervical cancer. SOX2, a pivotal regulator of stem cell pluripotency, has recently been implicated in the modulation of ferroptosis, a specialized form of iron-dependent cell death, in cancer dynamics. This study delineates the role of SOX2 in the ferroptotic landscape of cervical carcinoma.
Objective
To delineate the association between SOX2 expression and ferroptosis in cervical cancer and develop a robust, SOX2-centric model for predicting prognosis and enhancing personalized treatment.
Methods
A multidimensional approach integrating advanced bioinformatics, comprehensive molecular profiling, and state-of-the-art machine learning algorithms was employed to assess SOX2 expression patterns and their correlation with ferroptosis marker expression patterns in cervical cancer tissues. A prognostic model incorporating the expression levels of SOX2 and ferroptosis indicators was meticulously constructed.
Results
This investigation revealed a profound and intricate correlation between SOX2 expression and ferroptotic processes in cervical cancer, substantiated by robust molecular evidence. The developed predictive model based on SOX2 expression exhibited superior prognostic accuracy and may guide therapeutic decision-making.
Conclusion
This study underscores the critical role of SOX2 in orchestrating the ferroptosis pathway in cervical cancer and presents a novel prognostic framework. The SOX2-centric predictive model represents a significant advancement in prognosis evaluation, offering a gateway to personalized treatment for gynaecologic cancers.
Keywords: SOX2 Gene, Cervical Cancer, Ferroptosis, Bioinformatics
Introduction
Cervical cancer is a significant global health burden and is the fourth most common cancer among women worldwide (Sung et al. 2021; Cohen et al. 2019). It arises from the transformation of cervical squamous or glandular epithelial cells and is primarily attributed to persistent high-risk human papillomavirus (HPV) infections (Frevert and Taran 2022; Bogani et al. 2024; Golia D’Augè et al. 2024). Despite advancements in screening and prevention, the incidence and mortality of cervical cancer remain concerning, underscoring the urgent need for a comprehensive understanding of its molecular pathogenesis and the identification of novel therapeutic targets.
SRY-related HMG-Box 2 (SOX2) is a critical transcription factor that plays essential roles in stem cell maintenance, embryonic development, and tissue homeostasis (Yamanaka 2008; Takahashi and Yamanaka 2006). It belongs to the SRY-related HMG-box (SOX) family of transcription factors, and family members are characterized by a conserved high mobility group (HMG) domain. SOX2 has been implicated in various physiological and pathological processes, including cancer development and progression. Emerging evidence indicates that SOX2 is frequently dysregulated in different cancer types, contributing to tumour initiation, maintenance, and metastasis (Cho and Jung 2023).
Tumour stemness, a defining characteristic of cancer stem cells (CSCs), is associated with tumour initiation, therapeutic resistance, and disease relapse (Garcia-Ortega et al. 2023). SOX2 has emerged as a key regulator of tumour stemness, functioning as a master regulator of CSCs in various cancers (Zhang et al. 2023a; Afify et al. 2023; Martell et al. 2023; Guo et al. 2023). In cervical cancer, CSCs constitute a subpopulation of tumour cells with self-renewal capacity, clonogenic potential, and the ability to drive tumour growth (Cho and Jung 2023). Understanding the role of SOX2 in cervical cancer stemness is of paramount importance in deciphering the mechanisms underlying tumorigenesis and identifying potential therapeutic targets to combat CSC-driven tumour progression.
Iron, an essential element for cellular homeostasis, is involved in a plethora of cellular processes, including DNA replication, cell cycle regulation, and energy production (Wang et al. 2023). However, iron dysregulation can lead to the accumulation of excess intracellular iron, which generates reactive oxygen species (ROS) and triggers a nonapoptotic form of cell death termed ferroptosis (Stockwell 2022; Liang et al. 2019; Chen et al. 2021). Recently, accumulating evidence has suggested that iron metabolism and ferroptosis regulation play critical roles in tumour biology. Interestingly, SOX2 is associated with iron metabolism in neural stem cells (Chang et al. 2023), but its involvement in regulating iron homeostasis in cancers, including cervical cancer, remains largely unexplored.
This study aimed to investigate the functional role of SOX2 in cervical cancer, with a particular focus on its impact on tumour stemness and its potential regulation of ferroptosis processes. We explored the expression patterns of SOX2 in cervical cancer tissues and correlated its levels with those of CSC markers, clinicopathological parameters, and patient outcomes. Additionally, we analysed the associations between SOX2 expression and iron metabolism-related genes in cervical cancer cells, shedding light on the potential regulatory role of SOX2 in ferroptosis pathways.
A comprehensive understanding of the involvement of SOX2 in cervical cancer biology may provide novel insights into the mechanisms of tumorigenesis and therapeutic resistance. Furthermore, our study may reveal new avenues for targeted therapeutic interventions aimed at eradicating CSCs and modulating ferroptosis pathways to improve cervical cancer treatment outcomes.
Methods
Tumor stemness assay
We obtained RNA-sequencing expression data and clinical information for a cohort of 253 patients diagnosed with cervical cancer from the TCGA dataset, accessible through the portal at https://portal.gdc.com. Employing the OCLR algorithm, we computed the mRNAsi index as formulated by Malta et al. This index is grounded in the mRNA expression profiles, encompassing a total of 11,774 unique genes. Following the same analytical approach, we employed Spearman correlation on the RNA expression data. By subtracting the lowest value and subsequently dividing the outcome by the maximum value, we normalized the resulting dryness index within the [0, 1] range (Lian et al. 2019; Malta et al. 2018).
Human protein atlas (HPA) database
In this study, an investigation into the expression of SOX2 and its prognostic relevance in cervical squamous cell carcinoma and adenocarcinoma was conducted using the Human Protein Atlas (HPA) database. The HPA database is a valuable resource for exploring protein expression patterns and their associations with clinical outcomes (Kaminker and Timoshenko 2021).
SOX2 expression data: expression data for in cervical squamous cell carcinoma and adenocarcinoma tissues were retrieved from the HPA database. This included information on SOX2 expression levels, subcellular localization, and staining intensities.
Prognostic information: prognostic data related to cervical squamous cell carcinoma and adenocarcinoma, such as overall survival (OS) outcomes, were obtained from the HPA database.
The retrieved SOX2 expression data were analyzed to assess the differential expression of SOX2 in cervical squamous cell carcinoma and adenocarcinoma tissues compared to normal tissues. This analysis aimed to determine whether SOX2 expression exhibits significant alterations in these cancer types (Basha et al. 2018).
Prognosis analysis
We retrieved RNA-sequencing expression data as well as the relevant clinical data for individuals diagnosed with cervical cancer from the TCGA dataset, accessible via the URL (https://portal.gdc.com). The comparison of survival disparities among these cohorts was conducted utilizing the log-rank test. Moreover, to assess the prognostic efficacy of SOX2 mRNA, we employed the timeROC analysis (version 0.4) to measure and compare its predictive precision (Zhang et al. 2020; Lin et al. 2020).
Data on SOX2 gene expression and patient survival were retrieved from established databases (mention specific databases like PanCanSurvPlot (https://smuonco.shinyapps.io/PanCanSurvPlot/) and Kaplan–Meier plotter (https://kmplot.com/analysis/) relevant to CSCC). The datasets included information on patient demographics, clinical characteristics, treatment modalities, and follow-up outcomes (Lin et al.2022).
Survival analysis was conducted to determine the relationship between SOX2 expression levels and the survival outcomes of CSCC patients. The following metrics were evaluated: Overall Survival (OS), Progression-Free Survival (PFS), Disease-Specific Survival (DSS), Disease-Free Survival (DFS). Describe the statistical methods used to analyze the data. This might include Kaplan–Meier survival curves, log-rank tests for comparing survival distributions, and Cox proportional hazards regression to adjust for potential confounders and assess the independent effect of SOX2 expression on survival outcomes (Zhang et al. 2021).
Pan cancers analysis
We utilized the “exploration” module of the Tumor Immune Estimation Resource (TIMER, version 2.0) available at http://timer.cistrome.org/ to visually present the contrasting gene expression patterns of SOX2 across distinct tumor tissues and their respective normal counterparts (Han et al. 2023).
Differential genes expression and KEGG/GO analysis
We acquired RNA-sequencing expression profiles and corresponding clinical data for a cohort of 253 patients diagnosed with cervical cancer from the TCGA dataset, accessible via https://portal.gdc.com. Employing the limma package within the R software, we conducted an in-depth analysis of differentially expressed mRNAs. Our criteria for differential expression were set as “Adjusted P < 0.05 and Log2(Fold Change) > 1.5 or Log2(Fold Change) < − 1.5 (Han et al. 2023).”
To delve into the functional implications of potential targets, we conducted a comprehensive functional enrichment analysis. Gene Ontology (GO) emerged as a widely-utilized resource for annotating genes based on molecular function (MF), biological pathways (BP), and cellular components (CC). Furthermore, we harnessed the Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis, a valuable tool for comprehending gene functions and pertinent high-level genome functional insights (Yu et al. 2012).
For a more profound comprehension of mRNA's involvement in carcinogenesis, we harnessed the ClusterProfiler package (version: 3.18.0) within the R environment. This enabled us to dissect the GO functions of potential targets and to enrich the KEGG pathways. To illustrate our findings effectively, we employed the ggplot2 package in R to craft box plots, while heatmap visualization was facilitated using the pheatmap package within the R software (Yu et al. 2012).
Iron death assay
We procured RNA-sequencing expression profiles along with corresponding clinical data for a cohort of 253 individuals afflicted with cervical cancer. These valuable datasets were retrieved from the TCGA dataset, accessible via the URL https://portal.gdc.com. In order to explore the context of ferroptosis, we referred to the work by Ze-Xian Liu et al. titled “Systematic analysis of the aberrances and functional implications of ferroptosis in cancer,” to identify genes associated with ferroptosis.
Venn analysis and KEGG/GO enrichment
Venn diagram analysis: we executed Venn analysis to pinpoint shared genes between the sets of differentially expressed genes and iron death-related proteins (Yu et al. 2012).
Pathway and functional enrichment: the next step encompassed conducting an enrichment analysis on the intersecting gene pool. This was done to unearth KEGG pathways and GO terms that are intertwined with both SOX2 and iron death. To carry out this analysis, we utilized Metascape, a comprehensive resource accessible at (Wei et al. 2022).
Immune checkpoint analysis
We obtained RNA-sequencing expression data and the relevant clinical information for cases of cervical cancer from the TCGA dataset, accessible through the link https://portal.gdc.com. To ensure the robustness of immune score evaluations, we employed a tool called immuneeconv. This R software package amalgamates six cutting-edge algorithms, namely TIMER, xCell, MCP-counter, CIBERSORT, EPIC, and quanTIseq. Each of these algorithms has undergone rigorous benchmarking and possesses distinctive strengths (Yi et al. 2020; Ravi et al. 2018).
Relation analysis
We obtained RNA-sequencing expression profiles along with relevant clinical data for cervical cancer cases from the TCGA dataset, accessible at https://portal.gdc.com. Employing the R software GSVA package, we conducted an analysis using the ‘ssgsea’ method as the chosen parameter. To examine the relationship between genes and pathway scores, we further evaluated the Spearman correlation (Hanzelmann et al. 2013; Wei et al. 2020).
Gene set enrichment analysis (GSEA)
GSEA was conducted on CSCC gene expression data retrieved from The Cancer Genome Atlas (TCGA). Data preprocessing included normalization and cleaning. Differential expression analysis segregated samples into distinct groups, utilizing tools like DESeq2 or edgeR. GSEA, executed via GSEA software, involved gene sets from MSigDB or custom CSCC-specific sets, with parameters meticulously defined. Analysis outcomes included Normalized Enrichment Score (NES), False Discovery Rate (FDR), and P-values, identifying significant gene sets (FDR < 0.25, P-value < 0.05). Biological implications of these sets were examined, with potential validation through independent datasets or experimental methods, offering insights into CSCC's molecular underpinnings (Kar et al. 2017).
Sensitivity in drug
We forecasted the anticipated chemotherapeutic response for each individual sample utilizing data from the most expansive accessible pharmacogenomics repository, the Genomics of Drug Sensitivity in Cancer (GDSC), accessible at https://www.cancerrxgene.org/. This prediction procedure was executed via the R package “pRRophetic”. By employing ridge regression, we approximated the half-maximal inhibitory concentration (IC50) for the samples. All parameters were maintained at their default settings (Jiang et al. 2021).
To counteract potential batch effects, we integrated the application of the "combat" technique, while for the various tissue types, we accounted for their influences. Furthermore, in handling duplicate gene expression records, we summarized them using the mean value (Geeleher et al. 2014).
Prognostic analysis of ferrozois-associated genes
We obtained RNA-sequencing expression profiles and corresponding clinical data for cervical cancer patients from the TCGA dataset (https://portal.gdc.com). The data was processed by converting count data to TPM (Transcripts Per Million) and normalizing it using log2(TPM + 1). We retained samples with associated clinical information, resulting in cervical cancer samples for subsequent analysis.
To assess survival differences among groups, we used the Log-rank test. The predictive accuracy of ferrozois-associated genes and risk score was evaluated using time ROC analysis (v 0.4).
For feature selection, we employed the Least Absolute Shrinkage and Selection Operator (LASSO) regression algorithm with tenfold cross-validation, utilizing the R package glmnet (Hanzelmann et al. 2013).
We constructed a prognostic model using Multivariate Cox Regression analysis with the R package survival. The model was optimized using a multi-factor Cox regression followed by stepwise iteration.
Kaplan–Meier curves, P-values, and hazard ratios (HR) with 95% confidence intervals were generated using Log-rank tests and univariate Cox proportional hazards regression (Wei et al. 2020).
Construction of a predictive model
We retrieved RNA-sequencing expression profiles alongside corresponding clinical data for cervical cancers from the TCGA dataset, available at https://portal.gdc.com. Our analysis comprised univariate and multivariate Cox regression procedures, aimed at pinpointing the pertinent factors for constructing a reliable nomogram. To visually present the significance of each variable, encompassing P values, hazard ratios (HR), and 95% confidence intervals (CI), we employed the 'forestplot' R package.
Subsequently, leveraging the outcomes of the multivariate Cox proportional hazards analysis, we crafted a predictive nomogram. This nomogram served as a graphical tool that synthesized the various factors, which can be employed to gauge the risk of X-year overall recurrence for an individual patient. The ‘rms’ R package facilitated the integration of these risk factors into the nomogram, with each factor assigned points corresponding to its impact (Liu et al. 2020; Jeong et al. 2020).
Prognostic model evaluation
Tumor data and clinical details for cervical squamous cell carcinoma were retrieved from the TCGA database (https://portal.gdc.com), specifically focusing on datasets in STAR format. Following acquisition, TPM-formatted data were extracted and underwent a normalization process using the transformation log2(TPM + 1). Only those samples with comprehensive RNAseq and clinical data were selected for further investigative steps. The refined set of cervical squamous cell carcinoma samples was utilized in downstream analyses. The performance of various prognostic models was evaluated by examining their Decision Curve Analysis (DCA) curves, utilizing the ggDCA package within the R software framework (Yang 2022).
All the above analysis methods and R package were implemented by R foundation for statistical computing (2020) version 4.0.3.
Results
Expression of the sox2 gene across diverse cancers
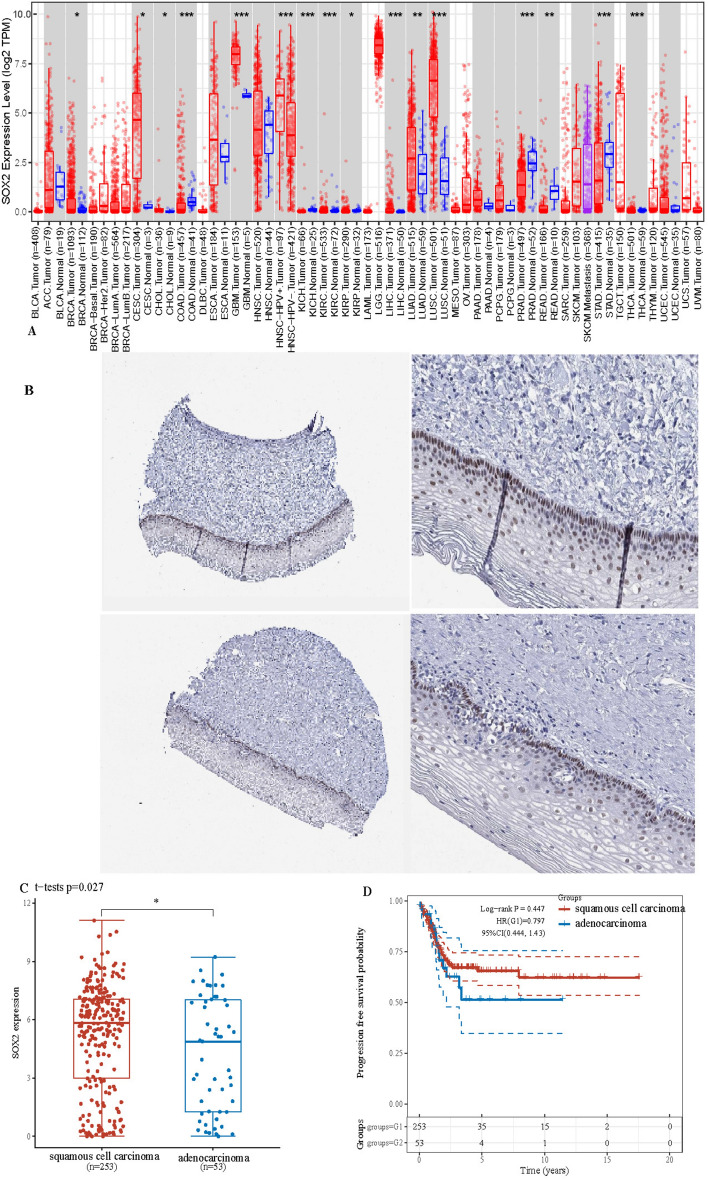
We initiated our investigation by examining the expression profile of the SOX2 gene across a spectrum of cancer types. An analysis of extensive datasets on cancer gene expression revealed significant upregulation of the SOX2 gene in squamous carcinoma tissues compared with their respective normal tissues (P < 0.001), such as cervical cancer (CESC), bile duct cancer (CHOL), and liver cancer (LIHC) tissues, as shown in Fig. 1A. In contrast, in adenocarcinoma, the expression of genes encoding SOX2 was notably downregulated (P < 0.01) in cancers such as stomach cancer (STAD), rectal cancer (READ), and prostate cancer (PRAD). These opposite SOX2 gene expression patterns suggest potential diverse roles in various cancers that may be linked to tumour type and progression. Our investigation extended to head and neck squamous cell carcinoma (HNSC), for which we made intriguing discoveries. In the initial analysis, the expression of the SOX2 gene in HNSC tumour tissues was not notably different from that in normal tissues. However, a marked difference was observed when we focused on the HPV status of HNSC patients. Among the HPV-positive HNSC samples, we observed a significant increase in SOX2 gene expression compared with that in their HPV-negative counterparts. This observation is consistent with our prior findings, reinforcing the notion that the SOX2 gene is highly expressed in squamous cell carcinoma.
Fig. 1.
SOX2 primarily acts in cervical squamous cells. A Demonstrates the expression of SOX2 across various types of solid tumors, indicating high expression in several tumors. B Depicts the distribution of SOX2 in cervical tissue, as revealed by the HPA database, primarily expressed in squamous epithelial cells. C Shows high expression of SOX2 in cervical squamous cell carcinoma. D In contrast, demonstrates lower expression of SOX2 in cervical adenocarcinoma. Additionally, compared to adenocarcinoma, cervical squamous cell carcinoma presents a more favorable prognosis. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
To gain insight into the interplay between the SOX2 gene and the subtypes of cervical cancer, we compared its expression patterns in squamous cell carcinoma and adenocarcinoma, as shown in Fig. 1B. The results revealed significant upregulation of the SOX2 gene in cervical squamous cell carcinoma, while its expression remained relatively low in cervical adenocarcinoma. These findings reinforce the notion that the SOX2 gene might be intricately linked to the development and distinct characteristics of different cancer subtypes.
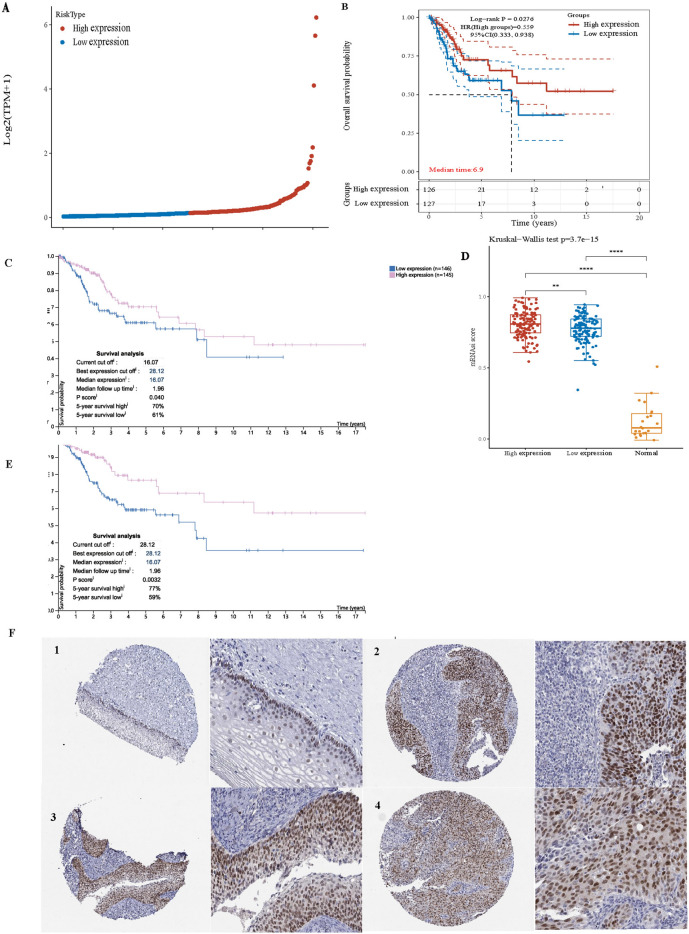
SOX2 is highly expressed in cervical squamous cell carcinoma. SOX2 expression is associated with the prognosis of cervical cancer.
In the TCGA dataset, we analysed the prognosis of cervical squamous cell carcinoma for patients grouped on the basis of high and low expression levels of SOX2. We found that the prognosis of the high-expression group was better than that of the low-expression group, as shown in Fig. 2A and B. Further investigation using the HPA dataset revealed a correlation between SOX2 expression and the prognosis of patients with cervical cancer. Patients with median to high SOX2 expression had a better prognosis than did those with low SOX2 expression, as depicted in Fig. 2C. Additionally, Fig. 2E shows that after the optimal cut-off value was determined, the difference became more pronounced. Considering that SOX2 is an important marker for tumour stemness, Fig. 2D indicates a higher stemness score in the high-expression group. Furthermore, the upregulation of SOX2 in cervical squamous cell carcinoma is evident from the HPA database, as shown in Fig. 2F.
Fig. 2.
Elevated levels of SOX2 expression in cervical cancer, with high SOX2 expression indicating a better prognostic outcome. A Significant variability in SOX2 expression across different cervical cancer samples. B Prognostic differences in cervical squamous cell carcinoma with varying levels of SOX2 expression, where the high-expression group has a better prognosis. C A substantial prognostic disparity in cervical cancer patients with different levels of SOX2 expression, again favoring the high-expression group. D Tumor stemness assays reveal that the high SOX2 expression group exhibits greater tumor stemness compared to the low-expression group and normal tissue. E After adjusting the appropriate CUT OFF value, the relationship between SOX2 expression levels and prognosis becomes clearer. F According to the HPA database, SOX2 expression in cervical cancer tissues (Figs. 2, 3, 4) is significantly higher than in normal tissue (Fig. 1). *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
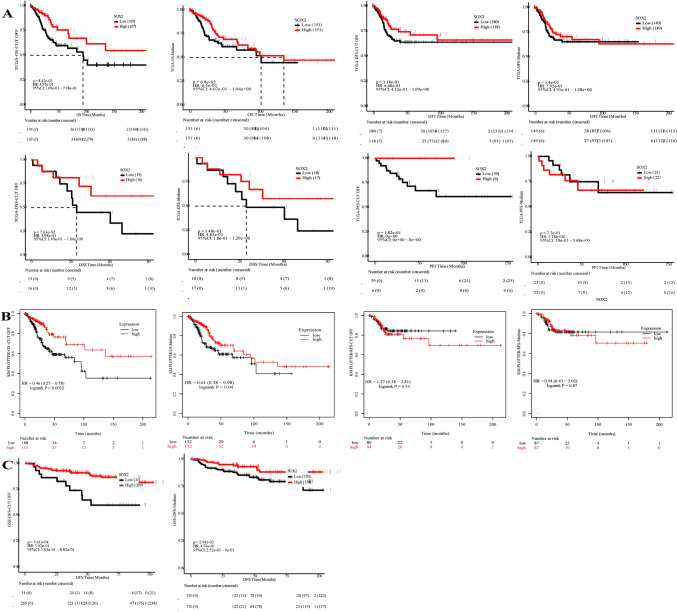
In multiple databases, SOX2 has been found to be correlated with various prognostic indicators in cervical squamous cell carcinoma.
Our focus is on the PanCanSurvPlot database and the Kaplan‒Meier plotter database. In these databases, cervical cancer samples can be grouped on the basis of median and CUTOFF values, and their clinical prognostic indicators can be evaluated.
As shown in Fig. 3A, in the PanCanSurvPlot database, the overall survival (OS) of cervical cancer patients was correlated with SOX2 expression levels, and high SOX2 expression was associated with a better prognosis. Similarly, Kaplan‒Meier plotter database analysis also supported the conclusion that higher SOX2 expression is associated with better OS (Fig. 3B). Furthermore, as shown in Fig. 3C, in the GSE44001 dataset, better disease-free survival (DFS) was observed in patients with high SOX2 expression.
Fig. 3.
Assessment of variations in SOX2 expression levels to evaluate the relationship between SOX2 expression and clinical prognosis. A Prognostic analysis of cervical cancer samples based on SOX2 median expression levels and optimal threshold levels reveals that SOX2 expression is associated with Overall Survival (OS) and Disease-Specific Survival (DSS) in cervical cancer patients, with higher SOX2 expression indicating better outcomes. B Data from the KMplotter database indicate a correlation between SOX2 and Overall Survival (OS) in patients with squamous cell carcinoma, with higher SOX2 expression suggesting a more favorable prognosis. C Findings from the GSE dataset reveal a relationship between SOX2 and Disease-Free Survival (DFS) in cervical cancer, with patients exhibiting higher SOX2 expression having a better DFS. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
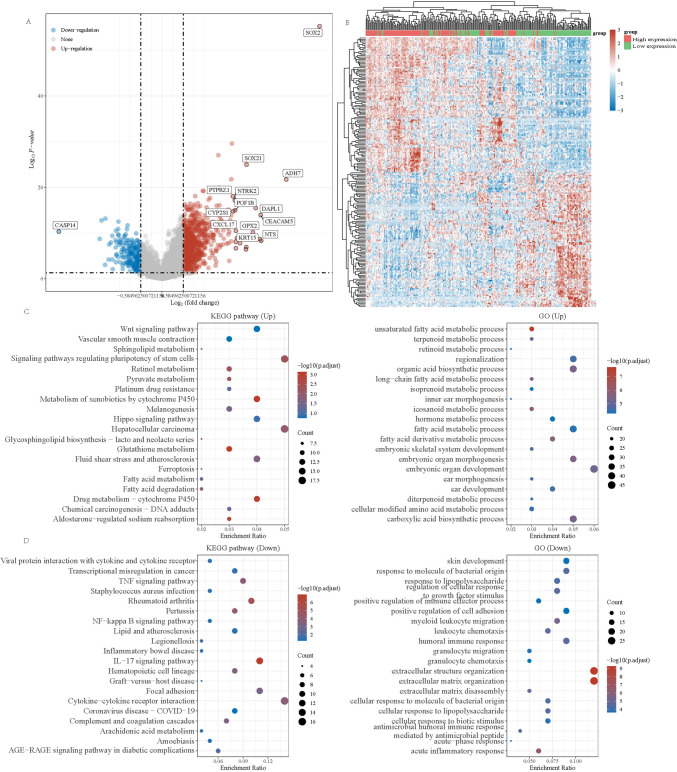
Stratification of sox2 gene expression and differential gene analysis
We further investigated the differentially expressed genes and their KEGG and GO analysis results in cervical cancer patients with high and low SOX2 expression. As shown in Fig. 4A and B, we constructed ial gene expression profiles and further analysed the KEGG and GO pathways, as depicted in Fig. 4C and D. In the KEGG analysis results, in addition to the WNT and HIPPO pathways, pathways related to ferroptosis were also enriched in the upregulated genes, while the main pathways enriched in downregulated genes were immune pathways and the TNF pathway.
Fig. 4.
Different SOX2 expression pattern and enrichment analysis in cervical squamous carcinoma in TCGA. A The volcano results show different gene expression in high and low SOX2 expression in TCGA. B The heatmap analysis. C KEGG pathway analysis of Upregulated genes. GOanalysis of upregulated genes. D KEGG pathway analysis of downregulated genes. GO analysis of downregulated genes. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
Moreover, the GO enrichment analysis of upregulated genes focused primarily on embryonic differentiation and tissue differentiation, whereas downregulated genes were enriched in extracellular matrix-related information.
Exploration of the association between sox2 gene expression and ferroptosis
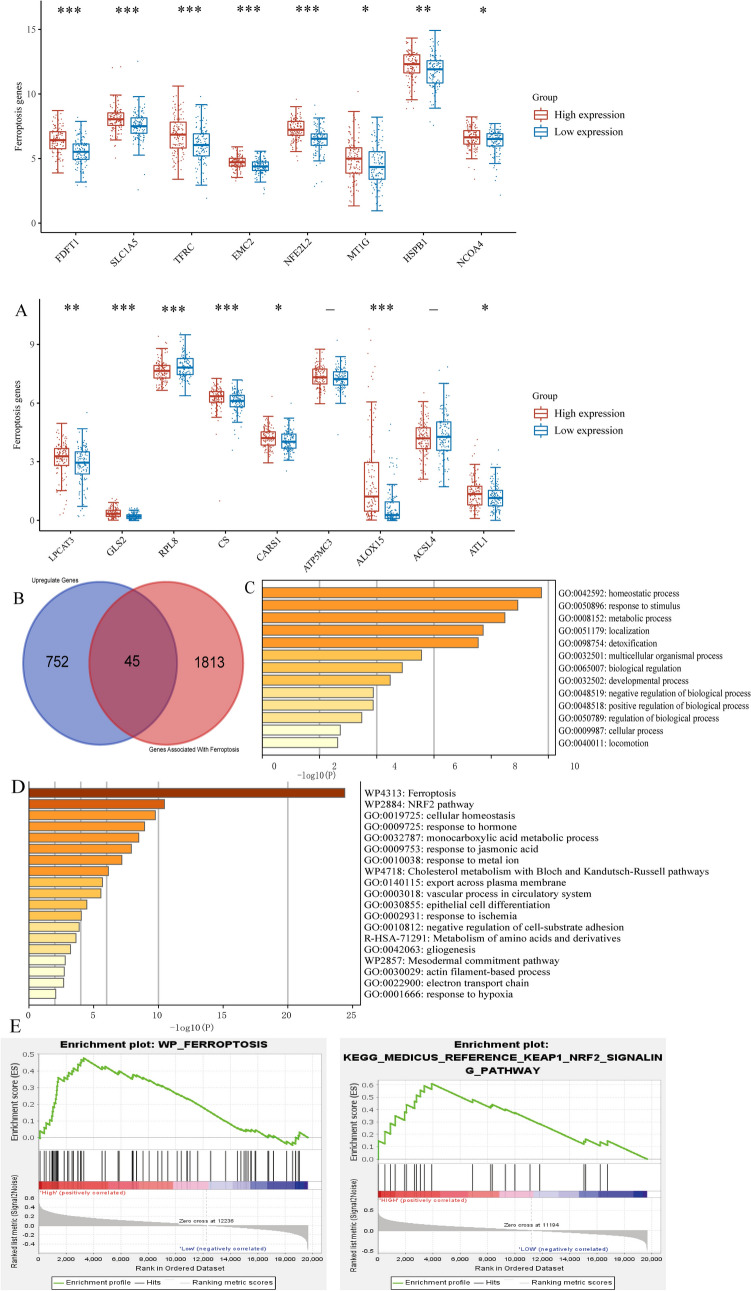
We further analysed the expression of genes related to ferroptosis in patients with high and low SOX2 expression, as shown in Fig. 5A. Various genes related to ferroptosis, such as TFRC and ALOX15, exhibited significant differential expression and were positively correlated with SOX2 expression. Furthermore, we intersected the upregulated genes from TCGA with ferroptosis-related genes to obtain a set of differentially expressed genes related to ferroptosis, as depicted in Fig. 5B.
Fig. 5.
Correlation between SOX2 expression levels and ferroptosis. A Key ferroptosis genes exhibit correlation with SOX2 expression. B Identification of 45 intersecting genes between upregulated differential genes and genes related to ferroptosis. C Conducting Gene Ontology (GO) analysis on the intersecting genes. D Performing Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on the intersecting genes, with the main findings involving ferroptosis and the NRF2 pathway. E Gene Set Enrichment Analysis (GSEA) results suggest a correlation between SOX2 expression and both ferroptosis and the NRF2 signaling pathway. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
Figure 5C and D further demonstrate through Metascape analysis that the KEGG and GO pathways related to ferroptosis and NRF2 were significantly enriched. Additionally, the GSEA results shown in Fig. 5E reveal a positive correlation between pathways related to ferroptosis and NRF2 and SOX2 expression, which is consistent with our previous findings.
Exploration of the relationship of the sox2 gene with immune features and checkpoint analysis
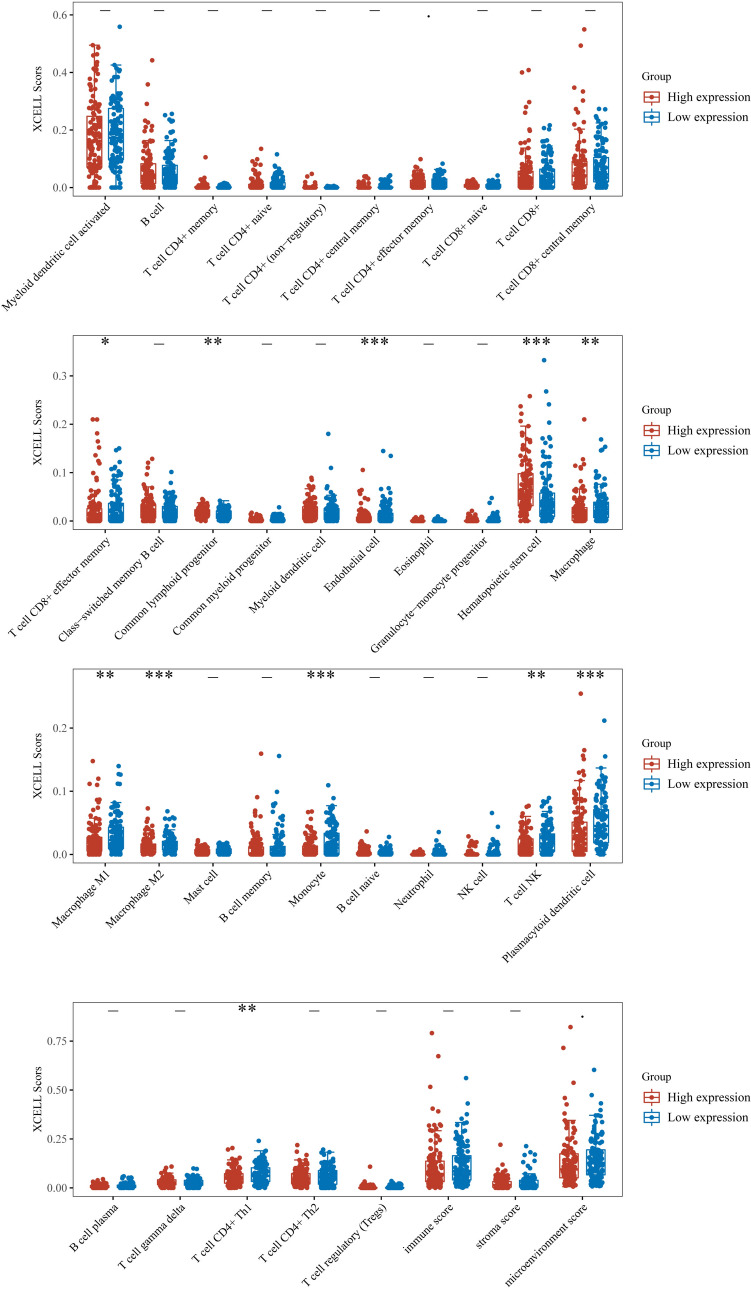
In our investigation, we expanded our exploration to encompass the immune features related to SOX2 gene expression in cervical squamous cell carcinoma, as shown in Fig. 6. Recognizing the crucial role of the immune system in tumour progression, we sought to reveal novel avenues for therapeutic interventions.
Fig. 6.
Correlation between SOX2 expression levels and immune cell infiltration. Assessment of the relationship between SOX2 and immune cells using the XCELL scoring method. The analysis reveals that, in various immune cells, the group with low SOX2 expression has a higher abundance of immune cells. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
To understand the potential immunomodulatory role of the SOX2 gene, we performed immune scoring on the basis of its expression levels. This analysis aimed to elucidate whether SOX2 gene expression is correlated with immune activation or suppression within the tumour microenvironment. Our findings could reveal its contribution to immune evasion mechanisms or immune surveillance enhancement.
Furthermore, we investigated the potential relationships between SOX2 gene expression and immune checkpoint genes. Immune checkpoints play pivotal roles in regulating immune responses against cancer cells. Examining the correlation between the SOX2 gene and these checkpoints might uncover additional layers of its involvement in shaping the tumour-immune interaction landscape. However, our findings suggest a limited association between the SOX2 gene and immune-related aspects.
Exploration of the correlation of the sox2 gene with pathways linked to cancer
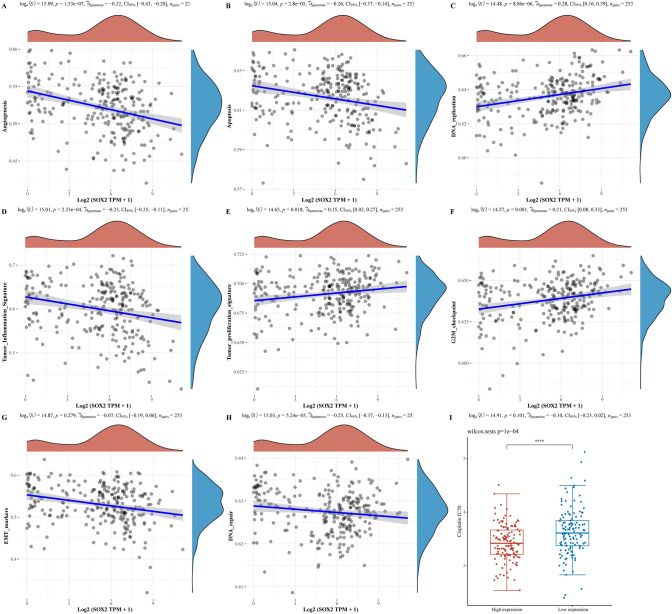
In our final endeavours, we comprehensively investigated the correlation between the SOX2 gene and the enrichment levels of pathways associated with cancer. This analysis aimed to reveal potential connections between SOX2 gene expression and key signalling pathways implicated in tumorigenesis. Our diligent exploration yielded intriguing results. We identified several pathways whose enrichment levels were significantly correlated with the expression of the SOX2 gene. These findings provide compelling evidence that the role of the SOX2 gene extends beyond its direct association with angiogenesis, the tumour inflammation signature, EMT and apoptosis, which are pathways relevant to tumorigenesis, as shown in Fig. 7.
Fig. 7.
The relationship between SOX2 and various tumor biology aspects and drug sensitivity results. A Angiogenesis is negatively correlated with SOX2 expression, indicating a possible inhibitory role of SOX2 in new blood vessel formation. B Apoptosis shows a negative correlation with SOX2 expression, suggesting that higher SOX2 levels may inhibit programmed cell death in tumors. C DNA repair is positively correlated with SOX2 expression, indicating a potential facilitative role of SOX2 in DNA repair mechanisms. D Tumor inflammatory characteristics are negatively correlated with SOX2 expression, suggesting that SOX2 may play a role in reducing inflammatory responses within tumors. E Tumor proliferation traits are positively correlated with SOX2 expression, indicating that SOX2 may promote tumor cell growth and replication. F. G2M checkpoint is positively correlated with SOX2 expression, suggesting a role for SOX2 in cell cycle regulation. G EMT (Epithelial-Mesenchymal Transition) markers show a potential negative correlation with SOX2 expression, indicating that SOX2 might be involved in the regulation of EMT processes. H DNA repair shows a negative correlation with SOX2 expression in a context different from part c, possibly suggesting complexity or context-dependent roles of SOX2 in DNA repair mechanisms. I Patients with high SOX2 expression in cervical cancer show higher sensitivity to platinum-based drugs, indicating a potential predictive role of SOX2 expression for chemotherapy response. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
Sensitivity of drugs according to sox2 expression
Furthermore, our study included drug sensitivity testing to explore the potential impact of SOX2 gene expression on treatment response. The analysis yielded a compelling observation: the group with higher SOX2 gene expression presented increased sensitivity to certain treatments, as shown in Fig. 7I. The heightened sensitivity of the high-SOX2 gene expression group in drug sensitivity testing has significant implications for therapeutic strategies. These findings suggest that targeting the gene SOX2 or its associated pathways may lead to improved treatment outcomes for patients with elevated gene SOX2 expression in the context of these specific treatments.
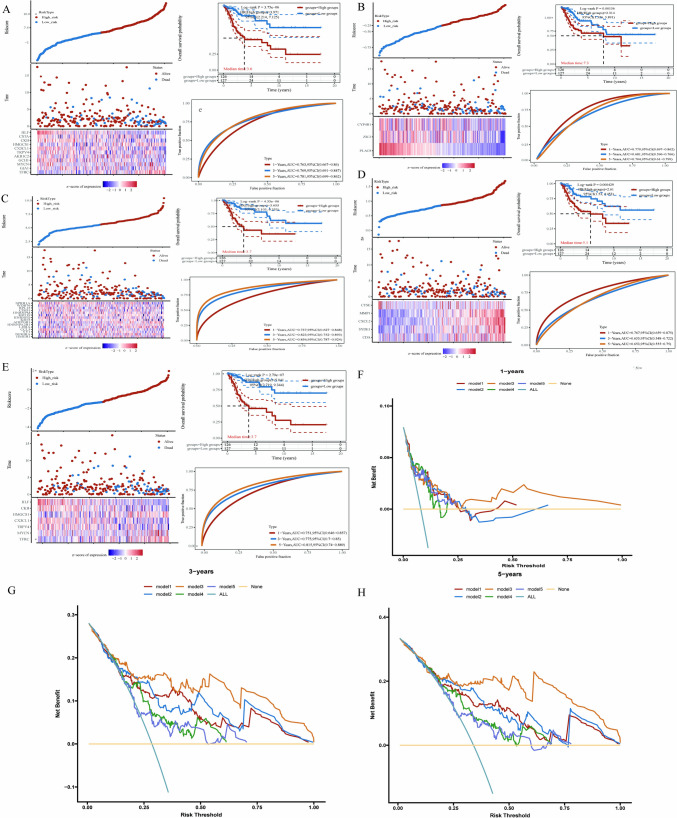
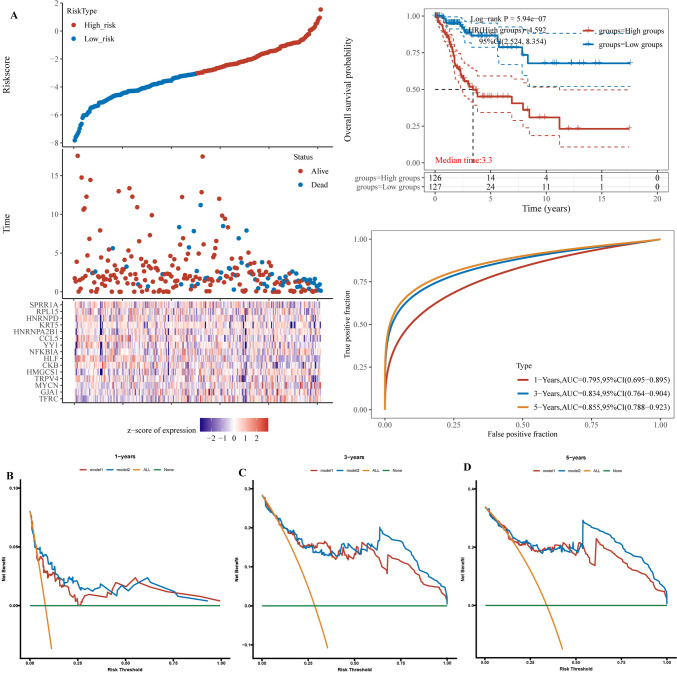
Construction and validation of a risk model based on to ferroptosis-associated genes in cervical squamous cell cancer patients
We constructed prognostic models using differentially expressed genes, as shown in Fig. 8A. We utilized the TCGA dataset and identified upregulated genes related to ferroptosis through LASSO Cox regression modelling to establish a prognostic model. We evaluated the prognostic model using the ROC curve and AUC value. As shown in Fig. 8B, we employed both the TCGA and GSE datasets to identify upregulated genes and constructed a prognostic model via LASSO Cox regression modelling, assessing its performance with the AUC curve. Similarly, as shown in Fig. 8C, we established a prognostic model using downregulated genes common to both the TCGA and GSE datasets through LASSO Cox regression modelling and evaluated its performance using the AUC value. As shown in Fig. 8D, we constructed a prognostic model using upregulated genes related to ferroptosis in the GSE dataset through LASSO Cox regression modelling and evaluated its performance via the area under the curve (AUC). Additionally, as shown in Fig. 8E, we employed the STEP model to establish a prognostic model for the dataset constructed in Fig. 8D and evaluated its performance using the AUC value.
Fig. 8.
Construction of various prognostic models and evaluation of their predictive performance. A Construction of a prognostic model based on upregulated genes and ferroptosis intersecting genes in the TCGA dataset. B Creation of a prognostic model using the upregulated genes common to both GSE and TCGA datasets. C Development of a prognostic model based on upregulated genes and ferroptosis intersecting genes in the GSE dataset. D Construction of a prognostic model using the downregulated genes common to both GSE and TCGA datasets. E Further analysis and simplification of model a to construct a streamlined prognostic model. F–H. Evaluation of 1-year, 3-year, and 5-year prognostic model performance, where model1 corresponds to model a, model2 to model e, model3 to model b, model4 to model d, and model5 to model c. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
Further evaluation of the models for the 1-year, 3-year, and 5-year data in Fig. 8F–H revealed that the model shown in Fig. 8E had the best performance.
Construction and evaluation of a prognostic model based on the common differentially expressed genes between the TCGA and GSE datasets
To construct a prognostic model centred on SOX2, we leveraged our existing dataset of differentially expressed genes. This dataset included genes that were commonly upregulated and downregulated in both the TCGA and GSE datasets, as well as the intersection of upregulated genes in the TCGA dataset with ferroptosis-associated genes and the intersection of upregulated genes in the GSE dataset with ferroptosis-associated genes. Prognostic models based on these gene sets were initially established via the least absolute shrinkage and selection operator (LASSO) regression method. To refine these models and mitigate overfitting, we applied a stepwise selection procedure (STEP) to optimize and construct more efficient prognostic models, as shown in Fig. 9A.
Fig. 9.
Construction and evaluation of a new prognostic model based on the common genes between TCGA intersecting with ferroptosis-related genes and GSE intersecting with ferroptosis-related genes. A Construction of a new prognostic model based on the common genes intersecting with ferroptosis-related genes from both the TCGA and GSE datasets. B Comparison of the 1-year prognostic performance of the new model with the prognostic model based on GSE intersecting with ferroptosis-related genes. C Comparison of the 3-year prognostic performance of the new model with the prognostic model based on GSE intersecting with ferroptosis-related genes. D Comparison of the 5-year prognostic performance of the new model with the prognostic model based on GSE intersecting with ferroptosis-related genes. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
Our analysis revealed that the prognostic accuracy of the intersecting ferroptosis-associated genes far surpassed that of the other gene sets. Moreover, decision curve analysis (DCA) of the prognostic models indicated that the predictive value of the intersecting genes between differentially expressed genes in the GSE dataset and ferroptosis-related genes was the highest. This finding underscores the potential of these intersecting genes as robust biomarkers for the prognostic assessment of cervical squamous cell carcinoma, particularly in relation to the SOX2 and ferroptosis pathways shown in Fig. 9B–D.
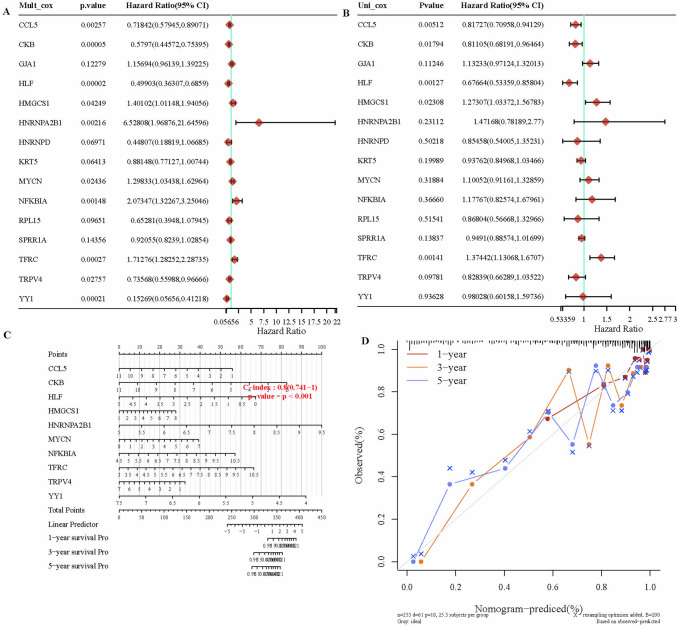
Analysis of the relevant genes in the final prognostic model
We evaluated the final prognostic model through univariate and multivariate Cox analyses of gene expression, with P values, hazard ratios (HRs), and confidence intervals presented in Fig. 10A–C. Nomograms predicting the 1-year, 3-year, and 5-year overall survival of cervical squamous patients were constructed, as depicted in Fig. 10D. The diagonal dashed line represents the ideal nomogram, whereas the blue, red, and orange lines represent the observed 1-year, 3-year, and 5-year survival data. We observed that the prognostic model achieved an AUC of 0.8, indicating its reliability and robustness.
Fig. 10.
Prognostic model analysis. A, B Univariate and multivariate Cox analyses of gene expression and clinical features, detailing the P-values, hazard ratios (HR), and confidence intervals. C Nomograms predicting the 1-year, 2-year, and 3-year overall survival of cervical cancer patients. D Calibration curves of the overall survival nomogram model in the discovery group. The diagonal dashed line represents the ideal nomogram, while the blue, red, and orange lines represent the observed 1-year, 2-year, and 3-year nomograms, respectively. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001
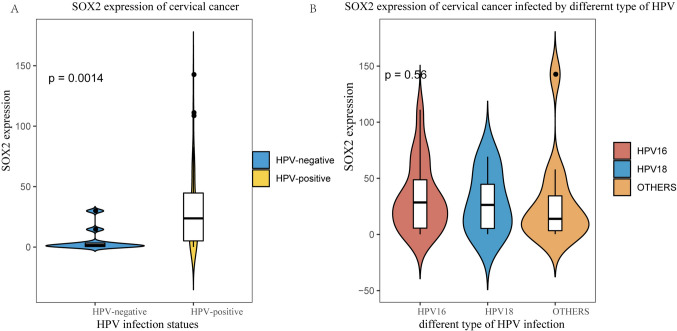
Sox2 expression in cervical cancer patients with different HPV infection statuses
To investigate the relationship between SOX2 expression and HPV infection in cervical cancer, we analyzed multiple GSE datasets. Our analysis revealed that SOX2 expression was significantly higher in HPV-positive patients compared to HPV-negative patients (Fig. 11A). This suggests that HPV infection is associated with an upregulation of SOX2 expression in cervical cancer.
Fig. 11.
SOX2 expression in cervical cancer based on HPV infection status. This violin plot A with a boxplot overlay shows the distribution of SOX2 expression in cervical cancer samples classified by HPV infection status. The blue area represents HPV-negative samples, while the yellow area represents HPV-positive samples. SOX2 expression is significantly higher in HPV-positive patients compared to HPV-negative ones. This violin plot B with a boxplot overlay demonstrates SOX2 expression levels in cervical cancer samples grouped by specific HPV infection types. The three groups represent HPV16 (red), HPV18 (blue), and other high-risk HPV types (orange). No significant differences in SOX2 expression were observed between these subgroups (p = 0.56)
Further analysis was performed to assess whether SOX2 expression varies among different types of high-risk HPV infections. HPV-positive patients were categorized into three subgroups: those infected with HPV16, HPV18, and other high-risk HPV types. Interestingly, no statistically significant differences in SOX2 expression were observed between these subgroups (Fig. 11B). These findings indicate that while HPV infection is associated with increased SOX2 expression, the specific type of high-risk HPV does not appear to significantly influence SOX2 expression in cervical cancer patients.
Discussion
The identification of distinct SOX2 gene expression patterns based on HPV status in HNSC highlights the potential interplay between viral factors and the tumour microenvironment (Leemans et al. 2018; The Cancer Genome Atlass Network 2015). Further investigations into the mechanistic link between HPV infection, SOX2 gene expression, and tumour progression could provide valuable insights into the aetiology of HPV-related squamous cell carcinoma (Salnikov et al. 2022).
In summary, our findings in the context of CESC further corroborate the association of the SOX2 gene with squamous cell carcinoma, emphasizing the multifaceted nature of its role in cancer development and its potential relevance as a therapeutic target.
Further analysis revealed an association between SOX2 gene expression and tumour differentiation. Strikingly, the high-expression group exhibited a substantial prevalence of squamous cell carcinoma, constituting the majority of tumours within this category. Conversely, the low-expression group had a significantly higher proportion of patients with adenocarcinoma, implying a close link between SOX2 gene expression and tumour differentiation. Our findings suggest that the SOX2 gene has diverse expression patterns across multiple cancer types, with potential implications for tumour differentiation and specific cancer subtypes. Notably, its elevated expression in cervical squamous cell carcinoma may contribute to its pathogenesis (Cho et al. 2022; Sedlic et al. 2020). These discoveries provide crucial leads for unravelling the functional mechanisms of the SOX2 gene in cancer.
The observed prognostic discrepancy might be attributed to various factors, including the intricate cross-talk between the SOX2 gene and other molecular pathways, as well as the unique immune landscape within cervical squamous cell carcinoma. Tumour heterogeneity and the differential contribution of the SOX2 gene to distinct cellular processes might also play pivotal roles in shaping the prognostic landscape.
The unexpected association between gene SOX2 expression and improved prognosis in cervical squamous cell carcinoma necessitates a comprehensive reevaluation of its functional role in this context. Further investigation into the underlying mechanisms governing the complex interplay between the SOX2 gene, tumour stemness, and prognosis will be crucial for advancing our comprehension of cervical squamous cell carcinoma biology and refining therapeutic strategies.
The results from our pathway and functional enrichment analyses underscore the complex interplay between gene SOX2 expression and the intricate biological processes within cervical squamous cell carcinoma. The enrichment of immune-related pathways and processes suggests a potential role for the gene SOX2 in modulating the immune microenvironment and tumour progression (Cao et al. 2023).
The novel connection between SOX2 gene expression and ferroptosis further expands the potential avenues for therapeutic intervention (Nie et al. 2023). Understanding how the SOX2 gene influences ferroptosis susceptibility in cervical squamous cell carcinoma cells may reveal novel strategies to sensitize tumours to ferroptosis-inducing therapies (Di Fiore et al. 2022).
Our comprehensive study not only reveals the enigmatic role of the SOX2 gene in cervical squamous cell carcinoma but also illuminates its potential involvement in ferroptosis regulation. The intricate interplay between the SOX2 gene, differential gene expression, and ferroptosis-related genes emphasizes the complexity of cancer biology. This knowledge may guide the development of targeted therapies aimed at exploiting these pathways for improved clinical outcomes.
The revealed correlations between the SOX2 gene and diverse cancer-related pathways open new avenues for future investigations. Exploring how the SOX2 gene interplays with these pathways and dissecting its precise molecular mechanisms could pave the way for novel therapeutic strategies targeting the broader network of factors implicated in tumorigenesis.
While these findings provide promising insights, they also raise several intriguing questions. The mechanisms underlying the relationship between SOX2 gene expression and treatment sensitivity warrant further investigation. Additionally, translating these discoveries into clinical applications could revolutionize treatment paradigms and improve patient outcomes.
The observed differences in drug sensitivity aligned with gene SOX2 expression levels emphasize the potential utility of precision medicine approaches (Yuan et al. 2023). Tailoring treatments on the basis of a patient's genetic profile, including gene SOX2 expression, could increase treatment efficacy and minimize adverse effects.
The construction of this predictive model has promising clinical implications. By amalgamating genetic and clinical variables, the model offers a comprehensive perspective on patient prognosis. The ability to predict outcomes via genetic signatures alongside clinical parameters is a powerful step towards personalized medicine (Zeng et al. 2023).
Our research revealed a clear correlation between the expression of SOX2 and the prognosis of cervical cancer. Furthermore, the prognostic model constructed on the basis of its differentially expressed genes had a significantly high AUC value, indicating the precise predictive value of the model. These findings indirectly validate the significant role of SOX2 in ferroptosis. Additionally, our further investigations demonstrated that the model based on SOX2 can serve as an effective prognostic tool for cervical squamous cell carcinoma patients and potentially indicates target for subsequent treatments (Zhao et al. 2022; Zhang et al. 2023b). This approach aligns with the shift towards personalized medicine and underscores the potential to improve patient outcomes through informed and targeted interventions.
Author contributions
All authors have contributed significantly to the conception, design, data analysis, and interpretation of this study.
Funding
This article received no financial support or funding.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval and consent to participate
All authors are aware of the manuscript's submission and consent to its publication.
Consent for publication
All authors have approved the submission and publication of this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Afify SM et al (2023) Cancer stem cells as the source of tumor associated myoepithelial cells in the tumor microenvironment developing ductal carcinoma in situ. Biomaterials 301:122249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha O et al (2018) The DifferentialNet database of differential protein–protein interactions in human tissues. Nucleic Acids Res 46(D1):D522–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogani G et al (2024) HPV persistence after cervical surgical excision of high-grade cervical lesions. Cancer Cytopathol 132(5):268–269 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlass Network (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nat 517(7536):576–82 [DOI] [PMC free article] [PubMed]
- Cao M et al (2023) Characterization of immature ovarian teratomas through single-cell transcriptome. Front Immunol 14:1131814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S et al (2023) Ferrodifferentiation regulates neurodevelopment via ROS generation. Sci China Life Sci. 10.1007/s11427-022-2297-y [DOI] [PubMed] [Google Scholar]
- Chen X et al (2021) Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol 18(5):280–296 [DOI] [PubMed] [Google Scholar]
- Cho HJ, Jung HJ (2023) Cyclophilin A inhibitors suppress proliferation and induce apoptosis of MKN45 gastric cancer stem-like cells by regulating CypA/CD147-mediated signaling pathway. Int J Mol Sci 24(5):4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH et al (2022) RKIP induction promotes tumor differentiation via SOX2 degradation in NF2-deficient conditions. Mol Cancer Res 20(3):412–424 [DOI] [PubMed] [Google Scholar]
- Cohen PA et al (2019) Cervical cancer. Lancet 393(10167):169–182 [DOI] [PubMed] [Google Scholar]
- Di Fiore R et al (2022) Cancer stem cells and their possible implications in cervical cancer: a short review. Int J Mol Sci 23(9):5167. https://www.mdpi.com/1422-0067/23/9/5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevert ML, Taran FA (2022) Human papillomavirus vaccines: a great leap forward. Case Rep Womens Health 33:e00388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ortega MB et al (2023) Interferon-alpha decreases cancer stem cell properties and modulates exosomes in malignant melanoma. Cancers (Basel) 15(14):3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P, Cox NJ, Huang RS (2014) Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol 15(3):R47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golia D’Augè T et al (2024) State of the art on HPV-related cervical lesions. Italian J Gynaecol Obstet 36(02):135–137 [Google Scholar]
- Guo L et al (2023) Targeting ITGB4/SOX2-driven lung cancer stem cells using proteasome inhibitors. iScience 26(8):107302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L et al (2023) CLDN5 identified as a biomarker for metastasis and immune infiltration in gastric cancer via pan-cancer analysis. Aging (Albany NY) 15(11):5032–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SH et al (2020) Nomogram for predicting gastric cancer recurrence using biomarker gene expression. Eur J Surg Oncol 46(1):195–201 [DOI] [PubMed] [Google Scholar]
- Jiang Q et al (2021) Establishment of an immune cell infiltration score to help predict the prognosis and chemotherapy responsiveness of gastric cancer patients. Front Oncol 11:650673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JD, Timoshenko AV (2021) Expression, regulation, and functions of the galectin-16 gene in human cells and tissues. Biomolecules 11(12):1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar SP et al (2017) Enrichment of putative PAX8 target genes at serous epithelial ovarian cancer susceptibility loci. Br J Cancer 116(4):524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans CR, Snijders P, Brakenhoff RH (2018) The molecular landscape of head and neck cancer. Nat Rev Cancer 18(5):269–282 [DOI] [PubMed] [Google Scholar]
- Lian H et al (2019) Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Mol Oncol 13(10):2227–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C et al (2019) Recent progress in ferroptosis inducers for cancer therapy. Adv Mater 31(51):e1904197 [DOI] [PubMed] [Google Scholar]
- Lin W et al (2020) Characterization of hypoxia signature to evaluate the tumor immune microenvironment and predict prognosis in glioma groups. Front Oncol 10:796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Yang H, Shi Y, Cheng Q, Liu Z, Zhang J, Luo P (2022) PanCanSurvPlot: A Large-scale Pancancer Survival Analysis Web Application. BioRxiv. 10.1101/2022.12.25.521884
- Liu Z et al (2020) A lncRNA prognostic signature associated with immune infiltration and tumour mutation burden in breast cancer. J Cell Mol Med 24(21):12444–12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta TM et al (2018) Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173(2):338-354.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell E et al (2023) Compensatory cross-talk between autophagy and glycolysis regulates senescence and stemness in heterogeneous glioblastoma tumor subpopulations. Acta Neuropathol Commun 11(1):110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J et al (2023) ASCL1-mediated ferroptosis resistance enhances the progress of castration-resistant prostate cancer to neurosecretory prostate cancer. Free Radic Biol Med 205:318–331 [DOI] [PubMed] [Google Scholar]
- Ravi R et al (2018) Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFbeta enhance the efficacy of cancer immunotherapy. Nat Commun 9(1):741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov M et al (2022) The HPV induced cancer resource (THInCR): a suite of tools for investigating HPV-dependent human carcinogenesis. mSphere 7(4):e0031722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlic F et al (2020) mitochondrial ROS induce partial dedifferentiation of human mesothelioma via upregulation of NANOG. Antioxid (Basel) 9(7):606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR (2022) Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell 185(14):2401–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676 [DOI] [PubMed] [Google Scholar]
- Wang X et al (2023) Mitochondrial carrier 1 (MTCH1) governs ferroptosis by triggering the FoxO1-GPX4 axis-mediated retrograde signaling in cervical cancer cells. Cell Death Dis 14(8):508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J et al (2020) Characterization of glycolysis-associated molecules in the tumor microenvironment revealed by pan-cancer tissues and lung cancer single cell data. Cancers (Basel) 12(7):1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z et al (2022) APOBEC3B is overexpressed in cervical cancer and promotes the proliferation of cervical cancer cells through apoptosis, cell cycle, and p53 pathway. Front Oncol 12:864889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S (2008) Pluripotency and nuclear reprogramming. Philos Trans R Soc Lond B Biol Sci 363(1500):2079–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L et al (2022) Integrative dissection of novel lactate metabolism-related signature in the tumor immune microenvironment and prognostic prediction in breast cancer. Front Oncol. 10.3389/fonc.2022.874731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L et al (2020) Comprehensive analysis of the PD-L1 and immune infiltrates of m(6)A RNA methylation regulators in head and neck squamous cell carcinoma. Mol Ther Nucleic Acids 21:299–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan SF et al (2023) IL-1RA promotes oral squamous cell carcinoma malignancy through mitochondrial metabolism-mediated EGFR/JNK/SOX2 pathway. J Transl Med 21(1):473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C et al (2023) Dissection of transcriptomic and epigenetic heterogeneity of grade 4 gliomas: implications for prognosis. Acta Neuropathol Commun 11(1):133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z et al (2020) Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma. Cancer Cell Int 20:27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F et al (2021) The antitriple negative breast cancer efficacy of spatholobus suberectus Dunn on ROS-induced noncanonical inflammasome pyroptotic pathway. Oxid Med Cell Longev 2021:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T et al (2023a) Identification of cervical cancer stem cells using single-cell transcriptomes of normal cervix, cervical premalignant lesions, and cervical cancer. EBioMedicine 92:104612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X et al (2023b) Compartmentalized activities of HMGCS1 control cervical cancer radiosensitivity. Cell Signal 101:110507 [DOI] [PubMed] [Google Scholar]
- Zhao MY et al (2022) Propofol augments paclitaxel-induced cervical cancer cell ferroptosis in vitro. Front Pharmacol 13:816432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.