Abstract
Biofilm formation is an important mechanism of survival and persistence for many bacterial pathogens. These multicellular communities contain subpopulations of cells that display metabolic and transcriptional diversity along with recalcitrance to antibiotics and host immune defenses. Here, we present an optimized bacterial single-cell RNA sequencing method, BaSSSh-seq, to study Staphylococcus aureus diversity during biofilm growth and transcriptional adaptations following immune cell exposure. BaSSSh-seq captures extensive transcriptional heterogeneity during biofilm compared to planktonic growth. We quantify and visualize transcriptional regulatory networks across heterogeneous biofilm subpopulations and identify gene sets that are associated with a trajectory from planktonic to biofilm growth. BaSSSh-seq also detects alterations in biofilm metabolism, stress response, and virulence induced by distinct immune cell populations. This work facilitates the exploration of biofilm dynamics at single-cell resolution, unlocking the potential for identifying biofilm adaptations to environmental signals and immune pressure.
Subject terms: Biofilms, Infection
Microbial biofilms contain subpopulations of cells that display metabolic and transcriptional diversity. Here, Korshoj & Kielian present an optimized bacterial single-cell RNA sequencing method and use it to study Staphylococcus aureus diversity during biofilm growth and transcriptional adaptations following immune cell exposure.
Introduction
Bacterial infections represent a pervasive clinical problem that is increasingly complicated by the emergence of multidrug-resistant (MDR) strains, recognized as one of the greatest threats to human health worldwide1–4. One successful bacterial pathogen typified by MDR is Staphylococcus aureus (S. aureus)5. While a commensal in nearly one-third of the human population, S. aureus is transmitted across both hospital and community settings as a leading cause of post-surgical infection, skin and soft tissue infection, bacteremia, endocarditis, osteomyelitis, and medical device-associated infection6. In addition to the large arsenal of immune evasion molecules and antibiotic resistance genes encoded by S. aureus, a hallmark of this pathogen is its propensity for biofilm formation6,7. Biofilm is a key mechanism for survival and persistence in the infected host, leading to significant morbidity and mortality not only for S. aureus, but also other MDR pathogens including Escherichia coli, Klebsiella pneumonia, and Pseudomonas aeruginosa8. It has been estimated that approximately 65% of nosocomial infections are associated with biofilm formation9. Encased in an extracellular matrix comprised of polysaccharides, proteins, and nucleic acids, the multicellular biofilm community is highly recalcitrant to antibiotics and the host immune system7–9. A combination of bulk transcriptomics, bacterial mutants, and fluorescent reporter strains have been employed to identify metabolically and transcriptionally diverse subpopulations of bacterial cells within biofilm that have differing roles in surface attachment, dispersal and dissemination, stress-response, host defense, and persistence7,8. Understanding these communities has been hindered by lack of a high-throughput method to simultaneously measure the complex and stochastic interactions between distinct bacterial subpopulations.
Single-cell RNA sequencing (scRNA-seq) is widely used for transcriptional profiling of eukaryotic cells within a heterogeneous sample10. It has been applied to assess immune response dynamics during bacterial infection, including biofilm, identifying transcriptional changes in leukocyte metabolism, reactive oxygen species (ROS) production, and inflammatory mediator signaling specific to each immune cell type11–15. However, the use of scRNA-seq has traditionally been limited in prokaryotes based on the short half-life and low abundance of mRNA, lack of polyadenylated transcripts, and complex cell wall characteristics16–18. As a result, bulk RNA-seq methods have primarily been used to study bacterial pathogens and biofilm communities. However, bulk methods fail to capture heterogeneity and underrepresented populations altogether. A single-cell approach is necessary for a complete transcriptional landscape of biofilm heterogeneity and how biofilm is affected in response to distinct immune pressures, a critical step towards identifying novel anti-biofilm strategies.
Only recently have bacterial scRNA-seq methodologies been described, each employing unique protocol variations with respective pros and cons19–27. One major area of variation between described methods is how individual cells are labeled with distinct oligonucleotide barcode sequences for identification, with methods broadly separating into plate- and microfluidics-based barcoding approaches. Plate-based systems have utilized standard 96- or 384-well plates that impose inherent limitations on cell numbers19,21,24, while microfluidics-based approaches permit acquisition of increased cell numbers but require adaptation of costly commercial instrumentation22,23,25,26. Another technique employed fluorescence-activated cell sorting for bacterial cell separation and identification, but yields were limited to a few hundred cells20,27. A second major area of methodological variation is RNA capture, with most approaches utilizing random hybridization or mRNA-targeted probes. The use of targeted mRNA probes requires prior knowledge of the genome and desired targets, effectively limiting the number of genes analyzed23, while random hybridization provides unbiased insights into all possible genes but results in an overabundance of rRNA reads (i.e. >90%)19,21,22,24,25. Initial studies with random RNA hybridization omitted rRNA depletion, whereas more recent reports successfully incorporated rRNA depletion with Cas9 or RNase H22,25,26. In all published bacterial scRNA-seq methods to date, studies were limited to planktonic organisms and focused on proof-of-concept feasibility. Several reports examined transcriptional changes between different planktonic growth states19,21,26, whereas others observed transcriptional variation in planktonic culture upon treatment with antibiotics or other stimuli22,23,25,26.
Here, we present an advanced method and application of bacterial scRNA-seq to explore the heterogeneity of complex biofilm communities and transcriptional adaptations in response to immune cell challenge. Our technique, termed BaSSSh-seq (bacterial scRNA-seq with split-pool barcoding, second strand synthesis, and subtractive hybridization), employs an optimized protocol for RNA capture from bacterial cells with low metabolic activity, as seen in biofilm7,8. BaSSSh-seq uses plate-based split-pool barcoding to label individual cells, without the need for sophisticated commercial equipment28–31. Random hexamers are used for unbiased RNA capture during barcoding. Additionally, second strand synthesis replaces the highly inefficient process of template switching to generate cDNA libraries32, and an enzyme-free rRNA depletion method based on subtractive hybridization is used to significantly reduce rRNA contamination33. Through reduced enzyme usage and rRNA contamination, costs are decreased while concurrently increasing sequencing depth. This concept is important for bacterial scRNA-seq given the inherent sparseness of cellular mRNA. We established that diversity can be captured from bacterial cells with low metabolic and transcriptional activity within biofilm and coupled this with innovative computational assessments for identifying transcriptional heterogeneity and dynamics.
We applied BaSSSh-seq to study unique transcriptional signatures that differentiate S. aureus biofilm from planktonic growth and how biofilm alters its transcriptional profile in response to immune pressure, elevating bacterial scRNA-seq from proof-of-concept demonstrations to address complex biological interactions. An initial comparison of biofilm vs. planktonic growth demonstrated the ability to capture transcriptional heterogeneity within biofilm and validated the BaSSSh-seq methodology through extensive consistency with literature and experimental observations. We then explored biofilm transcriptional alterations in response to immune pressure by applying BaSSSh-seq to biofilm after direct co-culture with three major leukocyte populations that have well-documented roles in S. aureus infection: macrophages (MΦs), neutrophils (PMNs), and granulocytic myeloid-derived suppressor cells (G-MDSCs)34–36. Within the transcriptionally diverse subpopulations of biofilm, differential responses to each leukocyte population were observed. We further developed an analytical pipeline using a combination of unique computational assessments and existing bioinformatics packages for an enhanced multi-level visualization of biofilm transcription. Through integration of iModulon analyses, we achieved a high-level assessment of transcriptional regulatory networks across biofilm subpopulations in addition to gene-level characterization37–39. Likewise, trajectory analysis was used to identify transcriptional dynamics between S. aureus growth states and activation upon immune pressure40. Together, BaSSSh-seq provides the opportunity for studying biofilm growth dynamics and interactions with the immune system at a new level of resolution, promoting enhanced understanding of biofilm pathogenesis and the potential for rational design of new therapeutic strategies.
Results
BaSSSh-seq enables bacterial scRNA-seq of biofilm and incorporates rRNA depletion
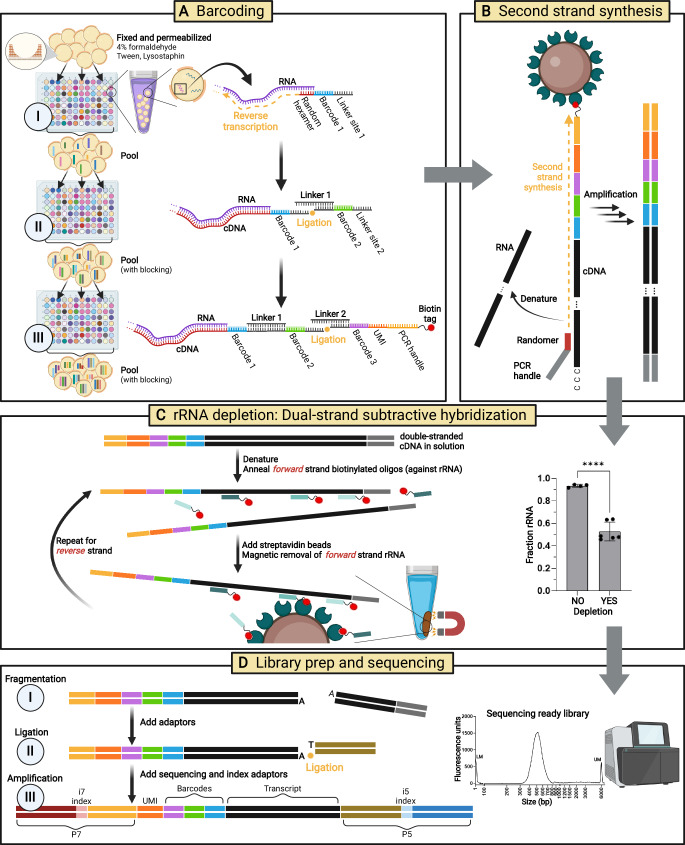
We employed split-pool barcoding to capture and label RNA transcripts (Fig. 1A, Supplementary Fig. 1), a technique originally described in eukaryotic cells and recently applied to prokaryotes19,21,28–31. Split-pool barcoding attaches a cell-specific combination of three oligonucleotide barcodes to RNA transcripts. Barcoding is performed in fixed and permeabilized bacteria (Supplementary Fig. 2) over three rounds consisting of an initial reverse transcription reaction where the RNA is captured, followed by two ligation reactions, interspersed with pooling and mixing steps. In our optimized implementation of split-pool barcoding, random hexamers were used for RNA capture during reverse transcription along with blocking unreacted barcodes with a set of complementary oligos during pooling to prevent non-specific and erroneous barcode ligations19. We also filtered, vortexed, and briefly sonicated cells between each barcoding step, which was previously shown to decrease the doublet rate21. Advantages of split-pool barcoding include its feasibility and cost, requiring only standard laboratory equipment. Other renditions of bacterial scRNA-seq have adapted commercial microfluidic instruments for cellular barcoding, requiring access to costly specialized equipment and reagents (Supplementary Fig. 1)22,23,25.
Fig. 1. BaSSSh-seq enables bacterial scRNA-seq of biofilm and incorporates rRNA depletion.
A Split-pool barcoding attaches a combination of three barcodes to intracellular RNA transcripts of fixed and permeabilized cells. The 5′ end of the terminal barcode oligo also includes a UMI, PCR handle, and biotin tag. B Following lysis, streptavidin magnetic beads are used to purify captured transcripts. Then double-stranded cDNA is synthesized via random primer second strand synthesis and PCR amplification. C Substantial rRNA depletion is performed using an enzyme-free dual-strand subtractive hybridization technique, where biotin-tagged oligos specific to 5S, 16S, and 23S rRNA fragments are annealed to each cDNA strand and magnetically removed with streptavidin beads. The rRNA content can be lowered from >90% to <50% (****, p-value < 0.0001 by unpaired t-test). Data includes 4 biological replicates with no depletion (25,000 cells per sample library, 1.5 × 105–2.5 × 105 paired-end reads per sample) and 6 biological replicates with depletion (120,000–150,000 cells per sample library, 2.4 × 107–4.4 × 107 paired-end reads per sample). Data are presented as mean ± standard deviation. D Libraries are constructed for Illumina sequencing through fragmentation, ligation, and amplification to generate constructs containing P5/P7 ends with unique i5/i7 index combinations. Schematic created in BioRender: Korshoj, L. (2024) BioRender.com/o39n335. Source data are provided as a Source Data file.
Following barcoding, cells were lysed, and captured transcripts were purified with streptavidin magnetic beads leveraging a biotin tag on the 5′-end of the terminal barcode oligo. Next, double-stranded cDNA was generated, where a second PCR handle was required for amplification. For this, we first attempted template switching based on an earlier plate-based barcoding method21, which exploits the terminal transferase activity of certain reverse transcriptase enzymes to anneal and synthesize the required PCR handle41. However, reliance on a short ~3-nucleotide sequence for annealing is highly inefficient and leads to significant transcript loss. Additional complications include concatamerization of the switching oligo when template concentrations are very low42, which we observed for bacterial biofilm RNA samples (Supplementary Fig. 3). We next tested the strategy of another plate-based barcoding method19 which used a second strand synthesis approach without subsequent amplification; however, we failed to generate any measurable cDNA libraries following this protocol. Therefore, we incorporated a random primed second strand synthesis step (Fig. 1B, Supplementary Fig. 4) as recently described for eukaryotic scRNA-seq, which significantly improved transcript yield compared to template switching32.
While random hexamer capture of RNA provides an unbiased survey of cellular transcripts, it leads to an overabundance of rRNA that can account for >90% of total sequencing reads, which we observed during protocol optimization and is consistent with known rRNA abundance in bacteria (Fig. 1C)19,21,22,33. Initial permutations of bacterial scRNA-seq omitted rRNA depletion, largely due to the difficulties in translating applicable depletion techniques from bulk RNA-seq to the in-cell reactions necessary for single-cell barcoding. Subsequent adaptations employed RNase H and Cas9 methods for rRNA depletion prior to barcoding, reducing levels by approximately 50%22,25. While a significant reduction, these procedures rely on additional enzymatic steps performed on fixed and permeabilized cells, potentially leading to substantial cell loss22, which we also observed in initial studies on cells prior to barcoding. When working with precious samples at low cell numbers, any losses can negatively impact or bias results. Therefore, we applied an enzyme-free rRNA depletion process to our double-stranded cDNA pool to reduce cell loss (Fig. 1C, Supplementary Fig. 5). This strategy uses subtractive hybridization, where short biotinylated oligos are annealed to rRNA-derived cDNA sequences and removed using magnetic beads33. Applied to double-stranded cDNA, the process is conducted on both forward and reverse strands. With this approach, rRNA levels were reduced to below 50% (Fig. 1C), consistent with other bacterial scRNA-seq methods but with the advantage of less cell loss from additional enzymatic steps.
After rRNA depletion, cDNA was fragmented to an optimal sequencing size (400–700 bp), ligated with short adaptors, and amplified to yield Illumina-compatible sequencing libraries containing P5 and P7 regions with dual indices (Fig. 1D, Supplementary Fig. 6). Library constructs contained a UMI for consolidating PCR duplicates and barcodes in read 2, and the transcript in read 1. Quality control measures throughout the BaSSSh-seq process are presented in the Methods and Supplementary Fig. 7. BaSSSh-seq was shown to faithfully capture bacterial transcriptomic profiles by comparing results with traditional bulk RNA-seq datasets of S. aureus biofilm previously generated in our laboratory that yielded statistically significant Pearson correlations (r = 0.570–0.581, p < 0.0001; Supplementary Fig. 8A–C)43. Given the intrinsic heterogeneity of biofilm, and variability in growth and sampling over the timescales between the two datasets, this bulk comparison provides strong validation of BaSSSh-seq fidelity while also highlighting the necessity of single-cell resolution (Supplementary Fig. 8D–E). For our S. aureus samples, BaSSSh-seq captured an average of 34 and 60 mRNA reads per cell during biofilm and planktonic growth, respectively (a range of 7 to >2000 reads per cell and 1 to >1000 detected genes), consistent with other bacterial scRNA-seq methods applied to S. aureus under planktonic growth conditions19,25. Considering that our analysis focused on biofilm, which is known to contain less metabolically and transcriptionally active cells7, this highlights the importance of our protocol improvements. Although the mRNA counts achieved for S. aureus biofilm are less than those reported for another widely studied Gram-positive pathogen, Bacillus subtilis, where 200–300 mRNA reads per cell were captured during planktonic growth21,23,26, this discrepancy can primarily be explained by its larger cell volume (~4–6×) compared to S. aureus44–46. Studies from eukaryotic scRNA-seq have established that low-coverage sequencing is sufficient to fully capture sample heterogeneity within a large number of cells47–50.
Biofilm growth is marked by extensive transcriptional heterogeneity and decreased metabolic gene expression at the single-cell level
Our first examination into S. aureus biofilm transcriptional complexity using BaSSSh-seq was with direct comparison to planktonic growth (Fig. 2A). As previously mentioned, earlier studies using bacterial scRNA-seq focused on planktonic growth and reported that unperturbed planktonic cultures are largely homogeneous19,21–23. Biofilm has been characterized to contain a heterogeneous population of cells with varying microstructural attributes that experience coordinated physiological changes throughout development51–55. Full mechanistic insights into signaling within the biofilm network remains elusive as our current understanding has principally relied on time-lapse microscopy with limited transcriptional reporter panels56–58. Comparisons between biofilm and planktonic growth have been explored in multiple studies over the past two decades with bulk transcriptomic or proteomic techniques59–62. Our BaSSSh-seq platform provides improved resolution for this growth condition comparison, and the potential for deeper mechanistic understanding of transcriptional signaling within diverse biofilm communities. Cells from biofilm and planktonic samples were collected simultaneously and fixed overnight before permeabilization the following day under identical conditions. Brightfield and confocal microscopy confirmed efficient biofilm disruption resulting in a single cell suspension for barcoding (Supplementary Fig. 2). For the first round of barcoding, cells from biofilm and planktonic cultures were processed separately so sample origin could be identified post-sequencing. Samples were combined for the second and third rounds of barcoding and processed together for sequencing. The planktonic culture yielded more cells with higher amounts of captured mRNA compared to biofilm (Supplementary Fig. 9A–C). This was expected based on the known differences in cellular activity during exponential growth of planktonic bacteria compared to biofilm where many organisms display a metabolically dormant phenotype21,62. To ensure an unbiased assessment with comparable cell numbers, sequenced cells for biofilm and planktonic samples were filtered at 7 and 28 non-rRNA reads per cell, respectively, which resulted in similar cell numbers (biofilm n = 3680 and planktonic n = 4231, Supplementary Fig. 9A) for downstream bioinformatic analysis. Importantly, differential expression analyses performed with unbalanced cell numbers have been shown to skew true positive and false positive identifications63, and we observed that any single cutoff equally applied to both biofilm and planktonic datasets was heavily unbalanced with 3–20× greater cell numbers for planktonic growth (Supplementary Fig. 10).
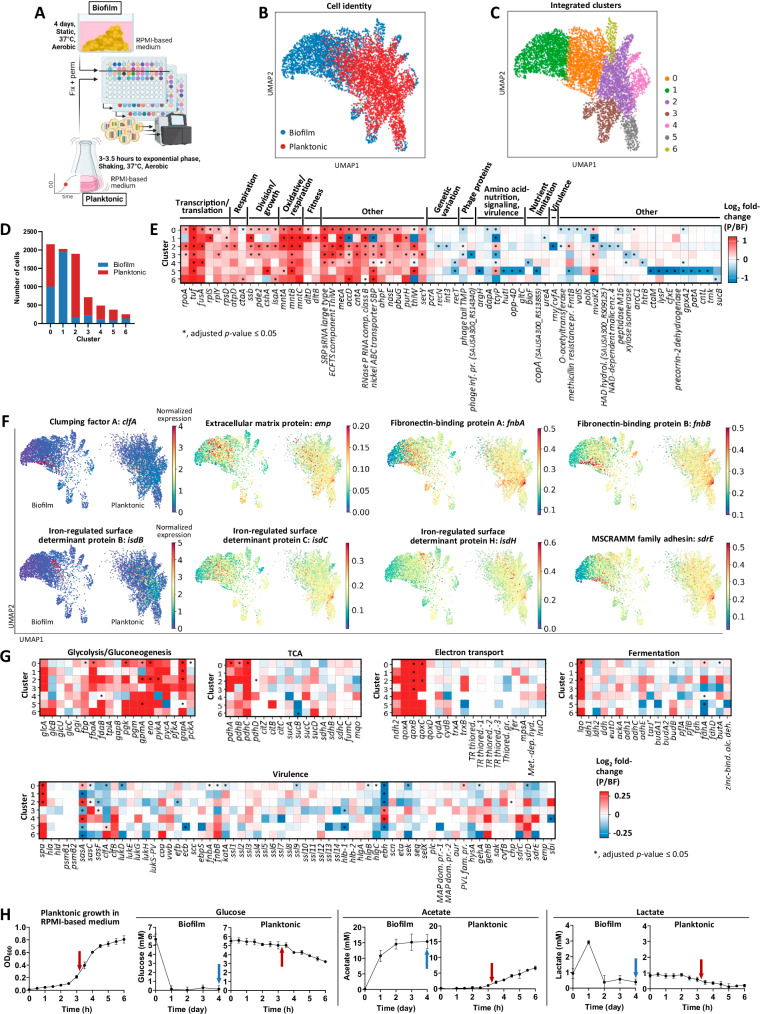
Fig. 2. Biofilm growth is marked by extensive transcriptional heterogeneity and decreased metabolic gene expression at the single-cell level.
A S. aureus biofilm was grown for 4 days under static conditions with daily medium replenishment, and planktonic culture was grown to exponential phase between 3 and 3.5 h with shaking at 250 rpm. Both biofilm and planktonic samples were grown in identical RPMI-based medium with aerobic incubation at 37 °C. Cells from biofilm and planktonic samples were fixed overnight before permeabilization under identical conditions. Cells from biofilm and planktonic cultures were separate for the first round of barcoding, then combined for the second and third rounds. The combined samples were processed through to sequencing. Schematic created in BioRender: Korshoj, L. (2024) BioRender.com/o01l597. B–C UMAP plots of the integrated biofilm and planktonic samples depicting (B) sample origin and (C) subpopulations identified with the Leiden algorithm. D Distribution of biofilm and planktonic cells across each cluster, reflecting 3680 cells for biofilm and 4231 cells for planktonic. E Marker genes specific to planktonic and biofilm growth across all clusters, represented as log2 fold-change of planktonic (P)/biofilm (BF). Red signifies upregulation in planktonic and blue signifies upregulation in biofilm. Additional genes are listed in Supplementary Data 1–7. False discovery rate (FDR)-adjusted p-values are noted. SRP signal recognition particle, ECFTS energy coupling factor transporter S, SBP substrate-binding protein, TPM tape measure protein. F Expression of exoproteome-associated genes overlaid on UMAP plots, separated by biofilm and planktonic conditions. Color represents normalized expression level on a per cell basis. G Comparisons of metabolic and virulence factor gene expression between planktonic and biofilm growth, represented as log2 fold-change of planktonic (P)/biofilm (BF). Red signifies upregulation in planktonic and blue signifies upregulation in biofilm. FDR-adjusted p-values are noted. TR thiol reductase, MAP MHC class II analog protein, TR thiored, SAUSA300_RS04260; TR thiored.−1, SAUSA300_RS04295; TR thiored.−2, SAUSA300_RS09235; TR thiored.−3, SAUSA300_RS13730; MAP dom. pr.−1, SAUSA300_RS10495; MAP dom. pr.−2, SAUSA300_RS10500. H Quantification of glucose, acetate, and lactate in culture supernatants collected during biofilm and planktonic growth (data are presented as mean ± standard deviation from 4 biological replicates). The planktonic growth curve in RPMI-based medium is also shown on the left (data are presented as mean ± standard deviation from 3 biological replicates). Arrows indicate the time of sample collection at which BaSSSh-seq was performed. Source data are provided as a Source Data file.
Clustering analysis using uniform manifold approximation and projection (UMAP) was performed on the integrated biofilm and planktonic datasets after batch balanced k nearest neighbors (BBKNN) alignment64. UMAP visualization revealed greater spatial distribution of biofilm cells, reflecting enhanced transcriptional diversity (Fig. 2B). In total, 7 transcriptionally unique subpopulations, or clusters, were identified across the integrated biofilm and planktonic dataset (Fig. 2C and Supplementary Fig. 9D). The largest cluster (cluster 0) was most equally distributed between biofilm and planktonic cells (Fig. 2D). The remaining clusters were largely biased towards biofilm or planktonic bacteria, reflecting the intrinsic transcriptional differences between the growth states. Differential gene expression was performed across all clusters using the MAST algorithm65. This hurdle model is important to account for the large number of dropouts, or non-detected genes in scRNA-seq datasets, which was observed with the biofilm sample given the low metabolic activity of some bacterial subpopulations. A known caveat of MAST is that log2 fold-change values can be small; therefore, minor differences cannot be disregarded as insignificant. Each cluster contained a set of marker genes for biofilm and planktonic growth, a subset of which are detailed in Fig. 2E (full lists in Supplementary Data 1–7). Overall, genes upregulated in the planktonic growth state were uniformly increased across the majority of clusters, whereas most upregulated genes within biofilm were unique to specific clusters, strongly highlighting its transcriptional heterogeneity.
During planktonic growth, several genes related to transcriptional and translational activity were more highly expressed compared to biofilm across most clusters (Fig. 2E). These include RNA polymerase subunit alpha (rpoA), elongation factors (tuf, fusA), and ribosomal proteins (rplS, rplY, rpsD)66–68. Upregulation of genes encoding an ATP synthase subunit (atpD) and primary heme A component of terminal oxidases (ctaA) suggest increased respiration in planktonic cells69,70. Heightened expression of genes for a single-stranded DNA-binding protein necessary for DNA replication (ssb), a regulator of secondary messenger cyclic di-adenosine monophosphate (pde2), a DEAD-box RNA helicase with known control over agr-mediated quorum sensing (cshA), and a housekeeping protein (isaA) are indicative of cellular division and environmental sensing associated with S. aureus exponential growth71–75. Planktonic cells also upregulated manganese transporters (mntABC) reported to combat oxidative stress generated from heightened respiratory activity during aerobic growth, and a set of lipoteichoic acid-associated genes for D-alanylation (dltD, dltB) that are linked to increased cellular fitness76–78.
As previously mentioned, upregulated genes within biofilm were primarily limited to specific clusters (Fig. 2E), reflecting increased heterogeneity. An interesting observation was that biofilm clusters were enriched for genes associated with genetic variation, including plasmid replication (pcrA), DNA repair (recN), integrase activity (int3), and recombinase activity (recT)79–84. Biofilm also expressed several phage protein genes (cataloged as ‘phage tail tape measure protein’ and ‘phage infection protein’). While likely remnants of previous phage insertion, phage activity has been postulated to promote bacterial persistence and survival during biofilm maturation and remodeling79. Upregulation of genes for arginine and lysine biosynthesis (argH and dapA), cysteine transport (tcyP), histidine metabolism (hutI), oligopeptide transport (opp-4D), and glutamate regulation (gltC) suggest reliance on amino acids for a range of cellular processes within biofilm since these genes are linked to nutrition, signaling, and virulence85–91. Further, nutrient limitation and stress were evident in biofilm by increased expression of genes for biotin synthesis (bioF), copper transport (copA), and urease (ureA) that is important for pH regulation within biofilm62,92,93. Increased RNase Y (rny/cvfA) levels were also seen in biofilm, which has been shown to tightly control mRNA expression for coordinated virulence gene activation94.
S. aureus exoproteome-associated genes have well-characterized roles in virulence and biofilm formation95–100. To examine their expression patterns across biofilm and planktonic growth states, gene expression was overlaid onto the UMAP space with the MAGIC imputation algorithm to remove noise obscuring underlying expression patterns, due to the inherent dropouts in scRNA-seq datasets101. Many exoproteome-associated genes displayed heighted expression in biofilm cells (Fig. 2F). Interestingly, some appeared more diffuse throughout biofilm (emp, fnbA, isdC), while others displayed more concentrated expression patterns within specific clusters (clfA, isdB, sdrE). The expression of these genes during planktonic growth was concentrated to a single cluster that exhibited widespread gene induction, which we show later to be the most transcriptionally active cells within the planktonic culture.
Next, metabolic and virulence factor gene expression was compared between S. aureus growth states. For metabolic assessments, differential expression was performed between planktonic and biofilm cells across all clusters for genes in major metabolic pathways including glycolysis/gluconeogenesis, tricarboxylic acid (TCA) cycle, electron transport, and fermentation (Fig. 2G). Glycolysis showed preferential upregulation in planktonic cells across all clusters compared to biofilm. Additionally, planktonic bacteria displayed increased expression of genes encoding the pyruvate dehydrogenase multienzyme complex (pdhABCD) that converts pyruvate to acetyl-CoA and acetate kinase (ackA) for acetate and ATP generation. This is consistent with known mechanisms of catabolite control protein A (CcpA) regulation under aerobic conditions and glucose availability, where the expression of glycolytic genes is increased while acetyl-CoA is converted to acetate and the TCA cycle is suppressed102. L-lactate-quinone oxidoreductase (lqo), which converts L-lactate to pyruvate for downstream ATP generation, was also upregulated in planktonic cells, suggestive of moderate lactate utilization for respiration and growth103–105. Planktonic cells further showed large induction of terminal oxidase genes (qoxABCD) required for respiration, consistent with the identified marker genes in Fig. 2E106. Expression of fermentative genes trended higher in biofilm, especially those related to formate metabolism (pflAB and fdh) that is important for biofilm structure and persistence43,107. These changes in metabolic gene expression aligned with extracellular glucose, acetate, and lactate concentrations in supernatants from biofilm and planktonic cultures as a validation of the BaSSSh-seq system (Fig. 2H). Compared to biofilm, exponential phase planktonic culture contained higher glucose levels that were progressively depleted, supporting the heightened expression of glycolytic genes. Acetate production in planktonic culture actively increased while the levels in mature biofilm plateaued, supporting the increased expression of pdhABCD and ackA in planktonic cells. Finally, lactate originating from the RPMI-based medium (Supplementary Fig. 9E) was actively consumed during planktonic growth whereas levels remained stable in the mature biofilm during later stage growth, supporting the observed upregulation of lqo. Virulence genes trended towards higher expression in biofilm compared to planktonic bacteria (Fig. 2G), with the exception of Protein A (spa)108. One of the most highly upregulated genes within biofilm was for a giant surface protein (ebh) with noted roles in regulating S. aureus clumping, virulence, osmolarity, surface attachment, and biofilm formation46,109. Another gene involved in surface attachment (sasA) was also increased in distinct biofilm clusters110. Collectively, this first comparison of biofilm and planktonic growth at the single-cell level illustrated the powerful capabilities of scRNA-seq to capture transcriptional heterogeneity within biofilm while at the same time validating the robustness of the BaSSSh-seq methodology through corroboration of observed transcriptional patterns with published literature.
Transcriptional regulation follows a trajectory from planktonic to biofilm growth
While the prior analyses uncovered alterations between biofilm and planktonic growth states at the gene expression level (Fig. 2), this did not provide insights into large-scale transcriptional regulation. Several computational tools exist for conducting pathway analysis for eukaryotic scRNA-seq, where a priori defined gene sets are assessed based on shared biological function111. Similar tools for prokaryotes are designed for higher density data produced by bulk RNA-seq and cannot be functionally translated to sparser single-cell datasets or contain an extraneous number of pathways confounding interpretation112,113. Recently, the concept of independent component analysis (ICA) has been applied to identify co-regulated, independently modulated gene sets (iModulons) within bacterial transcriptomes in an effort to understand the complex crosstalk between metabolism and gene regulation37–39. For S. aureus, ICA was applied to >300 bulk RNA-seq datasets across a range of conditions yielding 76 iModulons37. These iModulons were further condensed into 10 groups representing the transcriptional regulatory network. We integrated the ICA-determined gene sets from the S. aureus iModulonDB database into our BaSSSh-seq study as a new prokaryotic pathway analysis tool (Fig. 3A)39.
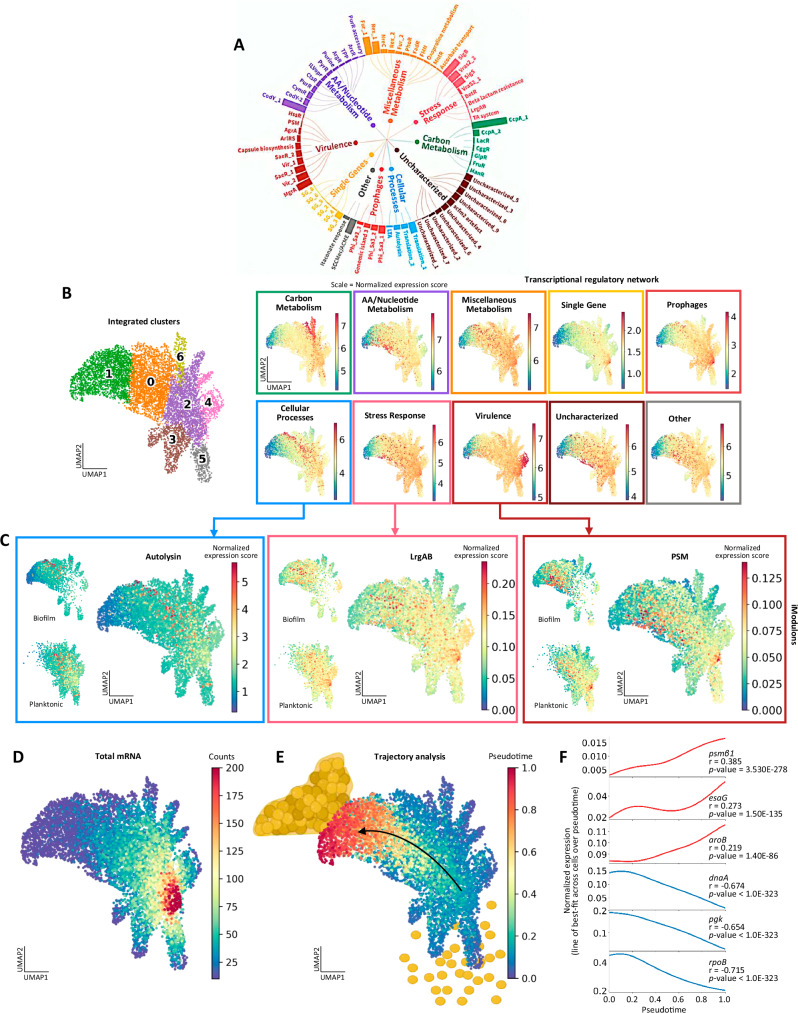
Fig. 3. Transcriptional regulation follows a trajectory from planktonic to biofilm growth.
A The S. aureus transcriptional regulatory network. Regulatory categories are comprised of iModulon sets, where each iModulon is comprised of an independently modulated set of genes. The size of the bars corresponds to the number of genes in the iModulon. B Transcriptional regulatory categories are overlaid on UMAP plots (right). Color represents normalized expression score per cell. For reference, the cluster identities (left) are shown as defined in Fig. 2C. C Autolysin, LrgAB, and PSM iModulon expression scores, linked to their respective transcriptional regulatory category. D Overlay of total mRNA counts on the UMAP plot, where color represents mRNA counts per cell. E Trajectory analysis (Palantir algorithm) identified a differentiation pathway through the integrated biofilm and planktonic samples, moving through pseudotime from planktonic to biofilm. F Subsets of genes display positive and negative correlation with the pseudotime trajectory, p-values are noted. Additional genes are listed in Supplementary Data 8–9. Source data are provided as a Source Data file.
Visualization of the S. aureus transcriptional regulatory network by overlaying expression scores onto the UMAP provided a meaningful coarse-grained view of metabolism, virulence, stress, and other cellular processes across biofilm and planktonic growth states (Fig. 3B, right). Activity within specific regulatory categories can be linked to previously defined clusters reflecting planktonic or biofilm growth (Fig. 3B, left, as defined in Fig. 2C). Overall, planktonic cells exhibited the highest expression scores for most regulatory categories concentrated in cluster 2, reflecting increased activity (Fig. 3B). In contrast, a region of cells in cluster 1 were inactive or dormant across all regulatory categories, which mainly represent biofilm. Individual iModulon expression scores were overlaid on the UMAP for a more granular view (Fig. 3C). The Autolysin, LrgAB, and PSM iModulons were selected based on the known associations of their respective genes with biofilm formation114,115. Interestingly, expression scores for these iModulons were increased in cells within biofilm but progressed seemingly along a path through the UMAP space from the most active planktonic cells in cluster 2 towards the most inactive biofilm cells in cluster 1, through the largest cluster 0.
To further explore this relationship, global transcriptional activity was visualized by overlaying total mRNA counts onto the UMAP (Fig. 3D). Planktonic cells, particularly within cluster 2 were most transcriptionally active, while biofilm cells in cluster 1 were least active. Trajectory analysis was then performed using the Palantir algorithm40. Adapted from eukaryotic scRNA-seq, trajectory algorithms identify paths of differentiation through a dataset at a single timepoint by quantifying divergences in gene expression between nearest-neighbor cells. Applied to our integrated biofilm and planktonic dataset, Palantir identified a trajectory from planktonic cells to biofilm, terminating at the most inactive or dormant cluster of biofilm cells (Fig. 3E and Supplementary Fig. 11A). The trajectory over pseudotime largely followed the patterns of iModulon expression (Fig. 3C). Further, a Pearson correlation analysis was conducted for gene expression over pseudotime to identify genes positively (Pearson correlation coefficient > 0) and negatively (Pearson correlation coefficient < 0) correlated with the trajectory (Fig. 3F). A subset of genes positively correlated with biofilm trajectory included phenol-soluble modulin (psmβ1), nuclease toxin system (esaG), and menaquinone biosynthesis (aroB)115–117. Genes negatively correlated with pseudotime were associated with replication initiation (dnaA), glycolysis (pgk), and transcription (rpoB)118–120. Comprehensive lists of positively and negatively correlated genes are provided in Supplementary Data 8–9. Furthermore, trajectory analysis was used to explore alternative transcriptional states in clusters 3, 4, 5, and 6 that radiate outward from cluster 2 on the UMAP (Fig. 2C). While each of these clusters showed moderate probability as an alternative branch of the overall trajectory early in pseudotime, the analysis converged to a terminal biofilm state represented by cluster 1 (Supplementary Fig. 11B), essentially reproducing earlier observations (Fig. 3E). This finding suggests these clusters likely represent intermediate states sharing transcriptional similarity to both biofilm and planktonic cells. The ability to quantify, visualize, and correlate transcriptional regulation across heterogeneous subpopulations of bacteria and growth trajectories provides a new level of resolution towards advancing our understanding of the mechanisms contributing to biofilm formation and persistence.
Biofilm shows coordinated transcriptional regulation
After comparing biofilm and planktonic growth and demonstrating the ability to visualize regulatory dynamics across heterogeneous bacterial populations and growth states, we next focused on a deeper analysis of biofilm heterogeneity and how this is altered in response to immune cell exposure. Our laboratory and others have studied the immune response to S. aureus biofilm infection in several animal models and humans34–36,121–125. The major leukocyte infiltrates associated with biofilm include MΦs, G-MDSCs, and PMNs, which are insufficient at clearing infection, which remains chronic34,35. These immune cell populations exhibit different metabolic, phagocytic, epigenetic, and transcriptional responses during S. aureus biofilm infection121–123. However, little is known about how biofilm adapts to each leukocyte type, especially at the single-cell level.
To investigate this question, BaSSSh-seq was applied to S. aureus biofilm directly co-cultured with MΦs, G-MDSCs, or PMNs (Fig. 4A). After co-culture, immune cells were lysed to prevent downstream RNA contamination. The bacterial cells from biofilm control (no immune cells) and the co-cultured biofilms were fixed overnight before permeabilization. Bacteria from each respective biofilm sample were barcoded separately for the first round, combined for the second and third rounds, and processed for sequencing. Sequencing reads were only aligned to the S. aureus transcriptome as a second means of preventing any potential contamination from eukaryotic RNA amplification. Sequenced bacteria were filtered at 15 non-rRNA reads per cell, which resulted in similar cell numbers per treatment (4500–6000 cells, Supplementary Fig. 12). UMAP clustering of the biofilm control yielded 7 transcriptional subpopulations (Fig. 4B). Independent UMAP clustering of co-cultured biofilms led to unique patterns (Fig. 4C and Supplementary Fig. 13), suggesting that S. aureus tailors its transcriptional response to each immune cell type, which is described in more detail below.
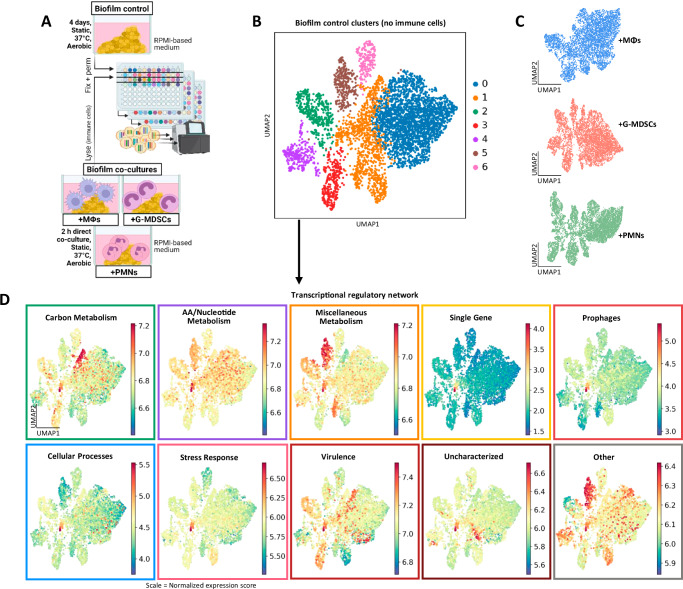
Fig. 4. Biofilm shows coordinated transcriptional regulation.
A S. aureus biofilm was grown for 4 days in RPMI-based medium under static, aerobic conditions at 37 °C with daily medium replenishment. After 4 days, biofilm was directly co-cultured with 5×105 mouse bone marrow-derived macrophages (MΦs), granulocytic myeloid-derived suppressor cells (G-MDSCs), or neutrophils (PMNs) for 2 h, whereupon immune cells were lysed with water. Cells from biofilm control (no immune cells) and the co-cultures were fixed overnight before permeabilization under identical conditions. Cells from each respective sample were separated for the first round of barcoding, then combined for the second and third rounds. The combined samples were processed through sequencing. Schematic created in BioRender: Korshoj, L. (2024) BioRender.com/z90k781. B UMAP plot of the biofilm control (n = 4655 cells), with colors denoting transcriptional subpopulations identified with the Leiden algorithm. C UMAP plots of independently clustered biofilms co-cultured with immune cells (n = 4544 cells for MΦ co-culture, n = 4780 cells for G-MDSC co-culture, n = 6125 cells for PMN co-culture). Responses to each immune population are explored in more detail in Fig. 6. D Transcriptional regulatory categories for the biofilm control are overlaid on UMAP plots. Categories correspond to the schematic in Fig. 3A. Color represents normalized expression score per cell.
We first characterized the biofilm itself to identify differences between the various transcriptional clusters using iModulon analyses (Fig. 4D). This revealed a complex picture of transcriptional regulation across biofilm subpopulations. Carbon metabolism and virulence regulatory networks showed strong coordination across the clusters, where expression scores were highest in clusters 2 and 3 while low in clusters 4, 5, and 6. Cluster 1 expressed a transcriptional signature that encompassed all the regulatory categories, and a stress response appeared to be moderately uniform across most biofilm clusters with a hotspot in cluster 1. Cluster 5 showed a prominent signature that was classified as miscellaneous metabolism (Fig. 4D). These initial high-level analyses help illustrate regulatory heterogeneity within biofilm, to aid in understanding the breadth of transcriptional diversity.
Biofilm subpopulations are characterized by diverse gene expression profiles
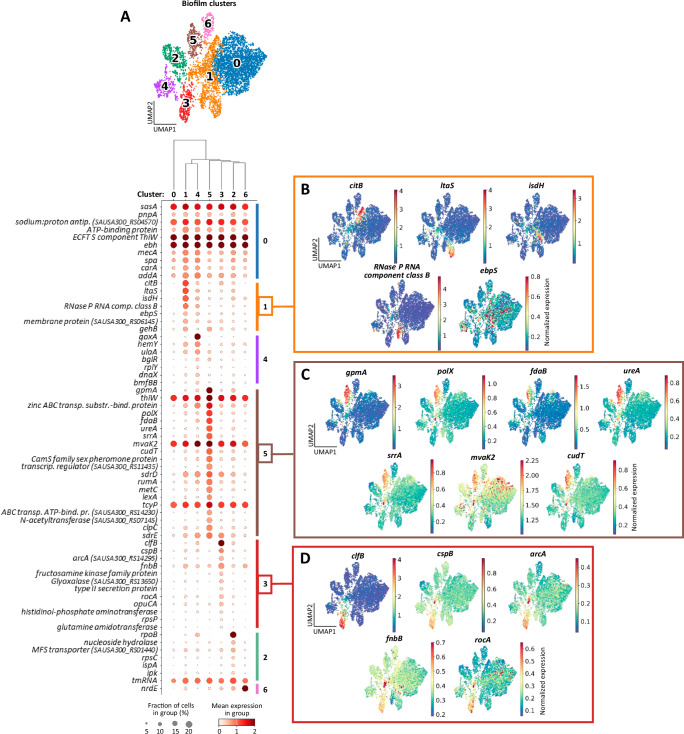
After obtaining an overview of biofilm transcriptional regulation with iModulon analyses, we next examined unique marker genes for each biofilm cluster to gain deeper insights into the functional state of each population, without influence from a planktonic comparison. Differential expression was performed for a given cluster vs. all others to identify marker genes (Fig. 5A). Of note, some biofilm clusters contained a greater number of statistically significant genes than others, and not all sets of marker genes translated to a meaningful classification. This may be influenced by clustering parameters, which were carefully considered to optimize unambiguous identification of unique marker gene sets (see Methods and Supplementary Fig. 13). The top 5 genes for each biofilm cluster are listed in Table 1.
Fig. 5. Biofilm subpopulations are characterized by diverse gene expression profiles.
A Dot plot with top marker genes per cluster from the biofilm control sample. All marker genes have FDR-adjusted p-value ≤ 0.05. The UMAP plot is shown above for reference as defined in Fig. 4B, and clusters are arranged in the plot according to transcriptional relationship, as illustrated in the dendrogram. A more detailed list of genes can be found in Table 1. ECFTS energy coupling factor transporter S, MFS major facilitator superfamily. Source data are provided as a Source Data file. B–D Marker gene expression overlaid on the UMAP plot for cluster 1 – n = 999 cells (B), cluster 5 – n = 272 cells (C), and cluster 3 – n = 288 cells (D). Color represents normalized expression per cell.
Table 1.
Marker genes from biofilm UMAP clusters, p-values are FDR-adjusted
| Cluster | Gene symbol | Locus tag | Product | TIGRFAM main role | TIGRFAM sub role | Log2 fold-change | Adj. p-value |
|---|---|---|---|---|---|---|---|
| 0 | sasA | SAUSA300_RS14395 | Serine-rich repeat glycoprotein adhesin SasA | Virulence | Virulence | 0.25 | 4.0E-15 |
| pnpA | SAUSA300_RS06315 | Polyribonucleotide nucleotidyltransferase | Transcription | Degradation of RNA | 0.24 | 4.0E-15 | |
| - | SAUSA300_RS04570 | Sodium:proton antiporter, TRAP transporter- DctM subunit | Transport and binding proteins | Carbohydrates, organic alcohols, and acids | 0.20 | 4.4E-12 | |
| - | SAUSA300_RS09575 | ATP-binding protein, DnaA regulatory inactivator Hda | DNA metabolism | DNA replication, recombination, and repair | 0.16 | 5.6E-06 | |
| - | SAUSA300_RS06415 | Energy coupling factor transporter S component ThiW | Biosynthesis of cofactors, prosthetic groups, and carriers; Transport and binding proteins | Thiamine; Other | 0.16 | 2.3E-08 | |
| 1 | citB | SAUSA300_RS06765 | Aconitate hydratase | Energy metabolism | TCA cycle | 1.33 | 8.3E-66 |
| ltaS | SAUSA300_RS03775 | Lipoteichoic acid synthase | Cell envelope | Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | 1.28 | 3.2E-65 | |
| isdH | SAUSA300_RS09160 | Iron-regulated surface determinant protein H | Transport and binding proteins | Cations and iron-carrying compounds | 1.11 | 1.3E-48 | |
| - | SAUSA300_RS15340 | RNase P RNA component class B | - | - | 1.07 | 7.0E-43 | |
| ebpS | SAUSA300_RS07485 | Elastin-binding protein EbpS | Virulence | Virulence | 0.35 | 1.3E-05 | |
| 2 | rpoB | SAUSA300_RS02820 | DNA-directed RNA polymerase subunit beta | Transcription | DNA-dependent RNA polymerase | 6.43 | <1.0E-323 |
| - | SAUSA300_RS01265 | Nucleoside hydrolase, translation initiation factor 2 gamma subunit | - | - | 0.45 | 3.8E-09 | |
| - | SAUSA300_RS01440 | MFS transporter | Cellular processes; Transport and binding proteins | Toxin production and resistance; Other | 0.34 | 3.9E-02 | |
| rpsC | SAUSA300_RS12120 | 30S ribosomal protein S3 | Protein synthesis | Ribosomal proteins: synthesis and modification | 0.30 | 1.1E-02 | |
| ispA | SAUSA300_RS08025 | Geranyltranstransferase | Biosynthesis of cofactors, prosthetic groups, and carriers | Menaquinone and ubiquinone | 0.29 | 6.1E-04 | |
| 3 | clfB | SAUSA300_RS14270 | Clumping factor B | Virulence | Virulence | 6.75 | <1.0E-323 |
| cspB | SAUSA300_RS14655 | Cold-shock protein | DNA metabolism; Cellular processes | DNA replication, recombination, and repair; Adaptations to atypical conditions | 0.75 | 2.0E-06 | |
| arcA | SAUSA300_RS14295 | Arginine deiminase | Energy metabolism | Amino acids and amines | 0.50 | 9.6E-11 | |
| fnbB | SAUSA300_RS13525 | Fibronectin-binding protein B | Virulence | Virulence | 0.47 | 2.4E-05 | |
| - | SAUSA300_RS14010 | Fructosamine kinase family protein | - | - | 0.45 | 9.0E-08 | |
| 4 | qoxA | SAUSA300_RS05175 | Quinol oxidase subunit 2 | Energy metabolism | Electron transport | 7.32 | <1.0E-323 |
| hemY | SAUSA300_RS09750 | Protoporphyrinogen oxidase | Biosynthesis of cofactors, prosthetic groups, and carriers | Heme, porphyrin, and cobalamin | 0.54 | 9.4E-03 | |
| ulaA | SAUSA300_RS01755 | PTS ascorbate transporter subunit IIC | - | - | 0.50 | 1.5E-02 | |
| bglR | SAUSA300_RS01375 | GntR family transcriptional regulator | Regulatory functions | DNA interactions | 0.38 | 7.3E-03 | |
| rplY | SAUSA300_RS02575 | 50S ribosomal protein L25/general stress protein Ctc | Protein synthesis | Ribosomal proteins: synthesis and modification | 0.36 | 7.3E-03 | |
| 5 | gpmA | SAUSA300_RS13045 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | - | - | 6.29 | <1.0E-323 |
| thiW | SAUSA300_RS05255 | ABC transporter ATP-binding protein | Transport and binding proteins | Unknown substrate | 2.28 | 6.5E-21 | |
| - | SAUSA300_RS12980 | Zinc ABC transporter substrate-binding protein | Transport and binding proteins | Unknown substrate | 1.72 | 1.1E-24 | |
| polX | SAUSA300_RS05610 | DNA polymerase/3’−5’ exonuclease PolX | DNA metabolism | DNA replication, recombination, and repair | 1.47 | 2.9E-26 | |
| fdaB | SAUSA300_RS14100 | Class I fructose-bisphosphate aldolase | Biosynthesis of cofactors, prosthetic groups, and carriers | Other | 1.46 | 2.4E-27 | |
| 6 | nrdE | SAUSA300_RS03845 | Ribonucleotide-diphosphate reductase subunit alpha | Purines, pyrimidines, nucleosides, and nucleotides | 2’-Deoxyribonucleotide metabolism | 6.97 | 6.8E-233 |
Cluster 1 (Fig. 5B) was identified as a transcriptionally active population, which was supported by ribonuclease P expression, an important enzyme in tRNA maturation126. This cluster was also associated with citB expression, suggesting an active TCA cycle under catabolite control protein E regulation during low glucose conditions present in biofilm (Fig. 2H)127. Other markers included lipoteichoic acid synthesis (ltaS), heme sequestration under iron limitation (isdH), and elastin binding/surface attachment (ebpS)128–130. Identifying cluster 1 as the most active population was corroborated by iModulon analyses (Fig. 4D), which showed modest expression scores throughout all regulatory categories. Cluster 5 (Fig. 5C) revealed a signature for alternative metabolism under stress. For example, gpmA and fdaB were enriched in cluster 5 and encode manganese-independent isozymes for two steps in glycolysis, suggesting metabolic activity under manganese limitation131,132. Elevated expression of the DNA polymerase gene polX indicates cells undergoing replication or repair133. Interestingly, PolX activity is manganese-dependent, suggesting that cells in cluster 5 could be prioritizing manganese usage under limiting conditions to maintain activity, although this remains speculative. Upregulation of the mevalonate pathway (mvaK2) further supports cellular activity in cluster 5134. Besides an increase in genes involved in limiting nutrient stress, the expression of ureA, srrA, and cudT reflect responses to acidic, nitrosative and hypoxic, and osmotic stress, respectively135–137, and were reflected in the iModulon network analyses (Fig. 4D). Biofilm cluster 3 (Fig. 5D) was enriched for genes involved in virulence and amino acid metabolism. For example, clumping factor B (clfB) and fibronectin-binding protein B (fnbB) are well-known virulence mechanisms of S. aureus involved in colonization and biofilm development96,97. The cspB gene codes for a cold-shock protein with implications in small colony variant formation that is a hallmark of biofilm cells138. Several genes involved in amino acid metabolism were increased in cluster 3, including arcA, which converts arginine to citrulline producing ATP, CO2, and NH3, as well as rocA which generates glutamate from proline139,140. Again, these classifications were supported by iModulon regulatory network analysis (Fig. 4D). Overall, these complex gene expression patterns support a dynamic and heterogeneous transcriptional profile in biofilm at the single-cell level.
Biofilm differentially responds to distinct immune pressures
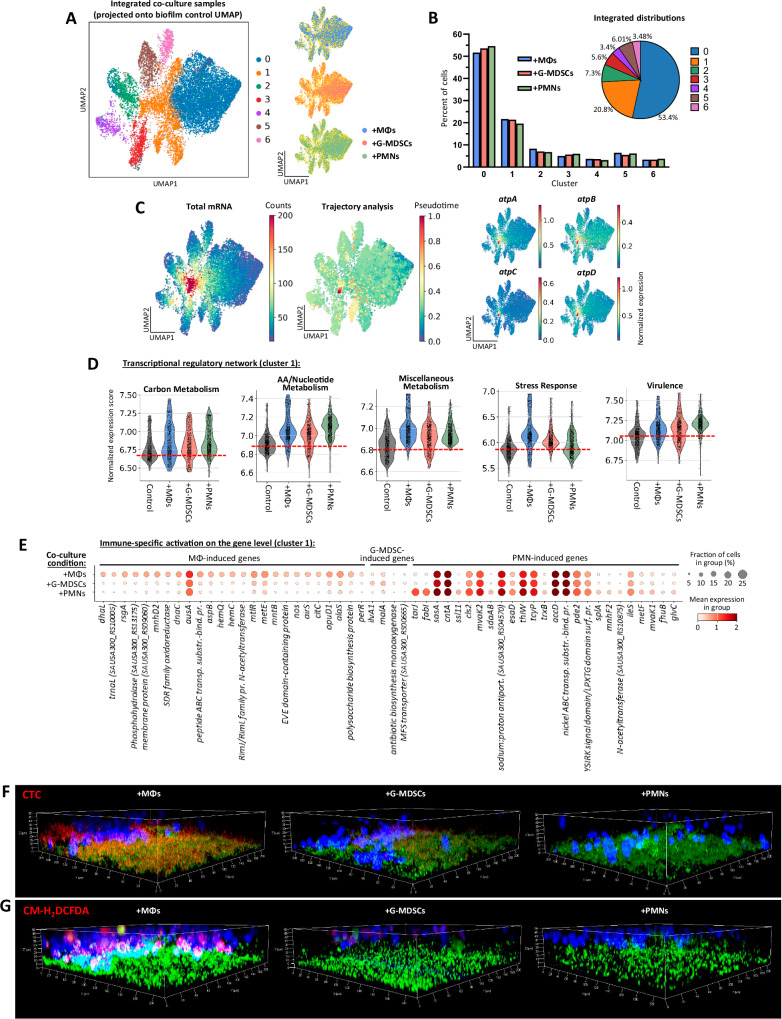
To explore how biofilm transcriptional profiles adapt to different immune pressures at single-cell resolution, we projected the bacterial cells from each biofilm co-culture condition (+MΦs, +G-MDSCs, and +PMNs) onto the biofilm control UMAP (Figs. 4 and 5). This provided a controlled basis for comparing each condition (Fig. 6A). All biofilms co-cultured with immune cells consistently mapped to the control UMAP with nearly equivalent distributions of bacterial cells across the different clusters, validating no bias in the system (Fig. 6B). As previously mentioned, biofilm cluster 1 contained the most transcriptionally active cells as depicted by overlaying total mRNA counts onto the UMAP space (Fig. 6C, left). Trajectory analysis also revealed a pseudotime convergence to cluster 1 (Fig. 6C, middle and Supplementary Fig. 11C) that corresponded with increased expression of ATP synthase genes (Fig. 6C, right), which are important for influencing immune cell activation and biofilm persistence69,141. Together, these findings suggest that the active biofilm population in cluster 1 experiences the most extensive transcriptional changes in response to immune pressure and was the focus of subsequent analysis. It is more challenging to compare less transcriptionally active clusters due to lower statistical power, which supports why cluster 1 was pursued.
Fig. 6. Biofilm differentially responds to distinct immune pressures.
A Integrated UMAP plot of biofilms co-cultured with MΦs, G-MDSCs, and PMNs. The biofilm control UMAP (Fig. 4B) was used as a template on which the co-culture samples were projected (using the ingest function of Scanpy). To the right, each individual co-culture condition is colored separately, with the other two co-culture conditions shown in yellow. B Bar plot showing the percentage of cells from each biofilm co-culture condition within each cluster. The pie chart shows combined cluster distributions for all co-culture samples. C (Left) Overlay of total mRNA counts on the integrated UMAP plot, with the highest number of captured transcripts present in cluster 1. (Middle) Trajectory analysis with the integrated biofilm-leukocyte co-culture samples (Palantir algorithm), which identified a differentiation pathway that converges to cluster 1 upon leukocyte exposure. (Right) Expression of atpA/B/C/D genes is concentrated in cluster 1, where the trajectory converges. D Transcriptional regulatory category expression within cluster 1 for biofilms co-cultured with immune cells compared to the biofilm control. Categories correspond to the schematic in Fig. 3A. Red dashed lines depict the average of the biofilm control for reference. E Top genes activated in biofilm cluster 1 in response to leukocyte exposure. Additional genes are listed in Supplementary Data 10–12. All genes have FDR-adjusted p-value ≤ 0.05 for differential expression (log2 fold-change) compared to the other co-culture conditions. F–G Evaluation of respiration and ROS activity using CTC (F) and CM-H2DCFDA (G) dyes (red) in biofilm (green)-leukocyte (blue) co-cultures with MΦs, G-MDSCs, or PMNs. Z-stack images were acquired (1 μm sections) and used to construct 3-D images. Color adjustments were applied uniformly across all images of the same experiment. Source data are provided as a Source Data file.
iModulon network activity across the different biofilm co-culture conditions revealed increased metabolic activity in cluster 1 in response to all three immune populations compared to the biofilm control, most prominently in utilization of miscellaneous amino acids and/or nucleotide sources (Fig. 6D). In contrast, stress response and virulence pathways were divergently regulated in response to the three immune populations. Specifically, a stronger stress response in biofilm cluster 1 was observed following MΦ exposure, whereas PMNs induced a heightened virulence response (Fig. 6D). G-MDSCs elicited the least perturbations in biofilm transcriptional profiles from the control. This was also confirmed by differential gene expression for each biofilm co-culture condition, where G-MDSCs induced minimal changes within biofilm (Fig. 6E, full gene lists in Supplementary Data 10–12) consistent with the iModulon network analysis and the known ability of G-MDSCs to promote S. aureus biofilm persistence by their anti-inflammatory activity34,35.
MΦ co-culture elicited the most unique differentially expressed genes in biofilm compared to G-MDSCs and PMNs (Fig. 6E). With regard to metabolism, MΦs upregulated dhaL that generates pyruvate from glycerol142. MΦ co-culture also induced evidence of a stringent response with increased rsgA and metE expression, which encode a ribosome-associated GTPase that inhibits translation upon sensing (p)ppGpp and a methionine biosynthesis gene tied to stringent conditions, respectively143,144. Genes for aspartate biosynthesis (aspB) and a non-ribosomal peptide synthase producing a protease inhibitor (ausA), both with links to virulence, were also induced in biofilm specifically following MΦ exposure145,146. However, the most upregulated genes during MΦ co-culture involved respiration and oxidative stress, where heme biosynthesis (hemC/Q) and nitric oxide synthase (nos) suggest active respiration under oxygen limiting conditions and oxidative stress elicited by MΦ activation147–149. Genes involved in mannitol metabolism (mtlR) and manganese acquisition and competition (mntB) also imply osmotic and redox pressure, with mntB suggesting superoxide dismutase activation150,151. Additional upregulated genes implicated in ROS detoxification include regulators of staphyloxanthin (airS) and peroxide resistance (perR), and a DNA helicase (dnaC) for replication and repair from oxidative damage152–154. Collectively, this suggests an adaptation to evade MΦ-mediated ROS production.
PMN co-culture also induced a unique transcriptional response in biofilm cluster 1 compared to MΦs and G-MDSCs (Fig. 6E). Upregulation of several metabolic genes related to amino acid catabolism (sdaAB) and methionine (metF) were observed144,155. Several respiration and oxidative stress genes were additionally increased, including staphylopine metal acquisition (cntA) and thioredoxin reductase (trxB)156,157. However, the most prominent genes upregulated during PMN co-culture are involved in cell wall maintenance and virulence, including tarJ, responsible for the rate-limiting step in CDP-ribitol synthesis for wall teichoic acids and fabI, a critical rate-limiting enoyl-ACP reductase for fatty acid synthesis158–160. Interestingly, the activities of both tarJ and fabI require NADPH oxidation to NADP+, indicating important regulation of cellular redox state. Additional upregulated genes involved in cell wall maintenance include mevalonate synthesis (mvak1/2), which affects both cell wall synthesis and membrane stabilization, and cardiolipin synthase (csl2)161,162. Increased cardiolipin synthase activity has been shown to inhibit PMN chemotaxis by reducing phosphatidylglycerol on the bacterial membrane, which is a chemoattractant163. Virulence genes induced in biofilm cluster 1 following PMN co-culture include a cell surface protein involved in surface attachment (sasA), a serine protease (splA), and superantigen-like protein 11 (ssl11)110,164,165. Similar to cardiolipin induction, SSL11 has been shown to arrest PMN motility by inducing adhesion without oxidative burst164.
Biofilm cluster 0 contained roughly 50% of the cells in the dataset. Transcriptional network analysis and quantification of total mRNA transcripts from cells in cluster 0 indicated that this was a metabolically and transcriptionally dormant population. Further studies into this group of cells could address the controversial issue of what defines a persister cell166,167. While these cells were not dead since the RNA would have degraded, their overall low activity suggests at least a portion may be persisters. Top marker genes for this cluster (Fig. 5A) included several surface proteins with known roles in adherence and virulence (sasA, ebh) and an exoribonuclease for RNA degradation (pnpA). Interestingly, cluster 0 exhibited evidence of ‘reawakening’ following immune cell exposure (Supplementary Fig. 14) reflected by a conserved upregulation of a RNA polymerase subunit previously shown to be correlated with a planktonic growth trajectory (rpoB, Fig. 3F), potentially suggesting reanimation to a more metabolically active population although this remains highly speculative.
The application of BaSSSh-seq to S. aureus biofilm co-cultures revealed the ability of biofilm to adapt and uniquely respond to distinct immune populations. Whereas anti-inflammatory G-MDSCs elicited minimal transcriptional changes, MΦs induced a prominent stress response to regulate respiration and oxidative damage, and PMNs induced genes related to cell wall maintenance and virulence. These observations were supported by confocal microscopy where S. aureus biofilms directly co-cultured with MΦs, G-MDSCs, or PMNs were stained with CTC (5-cyano-2,3-ditolyl tetrazolium chloride) or CM-H2DCFDA (6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate) to broadly probe respiration and ROS, respectively. Specifically, biofilm co-culture with MΦs led to increased respiration within biofilm (Fig. 6F) and the largest ROS signature (Fig. 6G) compared to G-MDSCs and PMNs, consistent with our transcriptomic analyses. Collectively, these findings demonstrate the sensitivity and selectivity of BaSSSh-seq to identify unique transcriptional alterations in S. aureus biofilm that can be used to examine how biofilm adapts transcriptional profiles in response to immune pressure.
Discussion
Here we present BaSSSh-seq, a bacterial scRNA-seq method incorporating a plate-based barcoding system with rRNA depletion. BaSSSh-seq was applied to study S. aureus biofilm heterogeneity and immune interactions, an advance from previous demonstrations of bacterial scRNA-seq on planktonic cells. This application captured vast transcriptional heterogeneity within biofilm compared to planktonic growth and permitted the detection of distinct biofilm responses tailored to different immune cell populations. In addition to the technical advances in scRNA-seq methodology, our analyses present a conceptual advance toward the understanding of complex biofilm communities by incorporating new computational pipelines that enable high-level regulatory network visualization and trajectory inference paired with gene-level expression quantification.
Our BaSSSh-seq methodology was validated by literature comparing alterations in gene expression and metabolism during biofilm vs. planktonic growth. Moreover, subsequent analyses laid the groundwork for exploration beyond simple validation. A current lack of understanding exists surrounding the intricately coordinated cellular networks that govern biofilm growth, stemming from inadequate high-throughput methods to measure the stochastic interactions between discrete subpopulations. A promising avenue for insights lies in the coupling of bacterial scRNA-seq with transcriptional regulation analysis, as implemented in our study. The iModulon-based assessments enabled cross-population relationships to be quantified and visualized. Furthermore, trajectory analysis provided another means to understand signaling dynamics, especially when linked to gene expression. While only a subset of genes correlating with the trajectory were discussed, many more remain unexplored (Supplementary Data 8–9). Several of these genes encode uncharacterized proteins that could potentially play key roles in biofilm formation and may represent attractive anti-biofilm therapeutic or prophylactic targets. An important future direction towards a better understanding of biofilm dynamics is to perform BaSSSh-seq during different stages of biofilm growth to assess 1) temporal alterations in gene expression; 2) changes in transcriptional regulatory networks through iModulons; and 3) clustering patterns during maturation. We did not detect many genes previously identified to be important during biofilm formation, such as the icaABCD and cidAB operons, which is likely because established biofilms were examined in this study57,60. Relating transcriptionally defined clusters to spatially defined microstructures and regions throughout the various stages of biofilm development would augment our understanding of biofilm growth and signaling, which could be achieved by constructing fluorescent reporters for genes that are enriched in distinct clusters.
BaSSSh-seq successfully generated powerful visualizations of biofilm transcriptional regulation paired with gene-level analyses for subpopulation characterization. The heterogeneity and coordinated patterns of gene regulation observed across biofilm clusters overwhelmingly illustrate how the ensemble-averaged expression from traditional bulk RNA-seq is insufficient. Accordingly, single-cell resolution also provides quantitative information on relative population sizes, a metric that is lost in bulk methods. Although many biofilm cells displayed a transcriptionally dormant phenotype (cluster 0), we focused our efforts on more active biofilm populations and how they interacted with the immune response. Our analyses demonstrated that biofilm undergoes dramatic transcriptional alterations that are tailored to the immune cell encountered. Although speculative, it is intriguing to consider that the most metabolically active biofilm clusters were responsive to MΦ and PMN challenge since these immune populations are major producers of ROS, RNS, and proteases that place strong pressures on bacteria123,148,152,153. In contrast, G-MDSCs do not exhibit antibacterial activity, so the biofilm does not need to expend substantial energetic resources to transcriptionally respond to this non-threat34,35. These findings have significant potential to inform more effective immunomodulatory therapies and support the concept of nutritional immunity described in the literature168. Future efforts will move in vivo, to explore the diversity of S. aureus adaptation and immune responses across different tissue niches.
Although highly functional, areas for improvement remain throughout the BaSSSh-seq methodology and analyses. For example, the number of barcoded cells with appreciable numbers of mRNA reads in biofilm samples was low. Insights from our comparisons of biofilm and planktonic cultures suggest this results from decreased transcriptional activity within biofilm. Nonetheless, membrane permeabilization conditions prior to barcoding could be more thoroughly studied to improve time and temperature for maximal barcode diffusion and RNA capture. Additionally, the barcoding could be expanded to 384-well plates to increase cell capacity by >60×. Sequencing depth also impacts the capture and detection of low-level transcripts, and with incorporation of rRNA depletion we improved cost efficiency and information content for sequencing runs, permitting usage of a mid-output kit on an Illumina NextSeq 500/550 series platform. However, availability of larger sequencers and kits exist for increasing sequencing depth >200×. As discussed further in the Methods, an inherent background noise exists, evident in the UMAP overlays in Figs. 2–5 where many genes were expressed at baseline levels throughout all clusters. This limitation restricted the statistical power of some analyses, and improvements would allow for higher confidence in identifying targets for experimental validation. Reduction in noise levels could be realized through adjustment of randomer concentrations in both reverse transcription and second strand synthesis steps, fragmentation conditions used in library prep, and/or modification of alignment parameters. Clustering itself could be further optimized to identify more meaningful classifications through further adjustments to parameter settings and/or future advances in clustering tools and algorithms. From a technical perspective, exploration of long-read sequencing presents a promising avenue that would allow fragmentation to be bypassed, leading to substantial noise reduction while potentially providing new insights into large-scale operon architecture. Several limitations are also evident from an experimental standpoint. First, as noted above, this study examined mature biofilm to assess how various immune cell subsets altered transcriptional programs. Performing BaSSSh-seq at regular intervals during biofilm development could provide new insights into fundamental populations that expand at key steps (i.e., attachment, exodus, and expansion)53. Second, spatial information about how specific biofilm transcriptional clusters relate to structural attributes (i.e., attachment, tower formation) is an interesting area to pursue as the resolution of spatial transcriptomic approaches improve. Based on the nature of this work describing BaSSSh-seq as a resource, the importance of specific S. aureus genes in biofilm biology or metabolism were not assessed, although we did validate changes in biofilm metabolism, respiration, and ROS as an initial step. Additionally, only one co-culture interval of biofilm and immune cells was examined (2 h) as a proof-of-concept for biofilm adaptation; however, the kinetics of these changes could be explored in future studies. Finally, in vitro biofilms grown in RPMI-based culture medium on coated plate surfaces do not replicate complex infection environments in vivo. While RPMI-based medium was necessary for leukocyte compatibility169–171, differences in glucose levels and other nutrients, as well as surface properties, are unable to model the full diversity of conditions encountered within the host. This further motivates the need to expand applications in vivo where differences in biofilm transcriptional profiles are expected in a niche-dependent manner based on nutrient availability and surface composition172.
Overall, the BaSSSh-seq method coupled with powerful computational approaches facilitates the high-throughput study of biofilm transcriptional heterogeneity at a new resolution. The datasets provide a rich resource for the biofilm community to explore, and the optimized protocols and analyses provide a mechanism to aid in identification of new therapeutic targets and strategies.
Methods
Oligos and reagents used throughout BaSSSh-seq are detailed in Supplementary Tables 1 and 2.
Bacterial strains
All sequencing experiments were performed with S. aureus USA300 LAC-13C173. For confocal microscopy, S. aureus GFP pCM29 and dsRed pVT1 expressing strains were used as previously described, with plasmids maintained during in vitro growth with 10 μg/mL chloramphenicol174,175.
RPMI-based medium
The RPMI-based medium used throughout all experiments for biofilm and planktonic cultures was RPMI-1640 supplemented with 10% heat-inactivated FBS, 1% L-glutamate, and 1% HEPES.
In vitro biofilm growth
24-well plates were coated overnight in 20% human plasma in 10X PBS at 4 °C to promote S. aureus attachment to initiate biofilm formation. Plasma coating solution was removed prior to seeding each well with 600 μL of S. aureus from an overnight culture grown for 16–18 h at 37 °C under aerobic conditions at 250 rpm using a 1:10 volume:flask ratio (25 mL RPMI-based medium in a 250 mL baffled flask) diluted 100× in RPMI-based medium. Plates were incubated under static, aerobic conditions at 37 °C for 4 days. Each day, 270 μL of spent medium was removed from each well, whereupon 300 μL fresh medium was carefully added to avoid disturbing the biofilm. For confocal microscopy experiments, biofilms were grown as described in 8-well chamber slides in 400 μL total liquid volume.
Planktonic growth
A single colony of S. aureus was inoculated into 25 mL RPMI-based medium at a 1:10 volume:flask ratio (250 mL baffled flask) for overnight (16–18 h) aerobic growth at 37 °C and shaking at 250 rpm. The following day, 250 μL of this overnight culture was inoculated into 25 mL fresh RPMI-based medium for outgrowth to exponential phase (3–3.5 h, OD600 = 0.35) under the same conditions as the overnight culture.
Overview of the BaSSSh-seq protocol for comparing biofilm and planktonic growth
Cells from biofilm and planktonic samples were collected, quickly mixed by pipetting, and vortexed before brief centrifugation at 12,000 × g for 1 min, and immediately resuspended in fixation buffer (4% formaldehyde in 1X PBS) for overnight incubation at 4 °C before permeabilization the next morning (both fixation and permeabilization performed in parallel and under identical conditions). Cells from biofilm and planktonic cultures were kept separate for the first round of barcoding, then combined for the second and third rounds. The combined samples were processed through second-strand synthesis, rRNA depletion, and library prep to sequencing. In total, ~200,000 cells were processed for sequencing from pooled and multiplexed individual libraries.
Preparation of primary MΦs, G-MDSCs, and PMNs
All immune cell types were prepared from the bone marrow of both male and female 8–10 week old C57BL/6J mice (RRID:IMSR_JAX:000664) as previously described176. The animal use protocol was approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee (#18-013-03). For MΦs, bone marrow cells were incubated in RPMI-based medium supplemented with 1X antibiotic/antimycotic solution (100 U/mL penicillin G, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin B), 50 μM 2-mercaptoethanol, and M-CSF from L929 cells for 7 days at 37 °C and 5% CO2, with medium changes on days 3 and 5. For G-MDSCs, bone marrow cells were incubated in RPMI-based medium supplemented with 1X antibiotic/antimycotic solution, 50 μM 2-mercaptoethanol, and 40 ng/mL each of G-CSF and GM-CSF for 4 days at 37 °C and 5% CO2, with 40 ng/mL of IL-6 added on day 3. After 4 days, G-MDSCs were purified with Anti-Ly6G MicroBeads. For PMNs, bone marrow was isolated and cells were immediately purified with Anti-Ly6G MicroBeads.
Overview of the BaSSSh-seq protocol for biofilm-leukocyte co-cultures
S. aureus biofilm was directly co-cultured with 5 × 105 primary MΦs, G-MDSCs, or PMNs for 2 h. After co-culture, all cells (bacteria and immune) were collected, quickly mixed by pipetting, and vortexed before brief centrifugation at 12,000 × g for 1 min. The cell mixture was resuspended in water for 10 min with brief, intermittent vortexing to preferentially lyse immune cells to prevent eukaryotic RNA contamination. After another centrifugation at 12,000 × g for 1 min, bacterial cells were immediately resuspended in fixation buffer (4% formaldehyde in 1X PBS) for overnight incubation at 4 °C before permeabilization the next morning. Cells from the biofilm control (no immune cells) and co-cultures were fixed and permeabilized in parallel under identical conditions. Cells from each respective sample were separated for the first round of barcoding, then combined for the second and third rounds. The combined samples were processed through second-strand synthesis, rRNA depletion, and library prep to sequencing. In total, ~400,000 cells were processed for sequencing from pooled and multiplexed individual libraries.
Confocal microscopy
Biofilms were grown in 8-well glass chamber slides and visualized during immune cell co-cultures using confocal laser scanning microscopy (Zeiss 710) with a 40× oil lens. Z-stack images were acquired (1 μm sections) and used to construct 3-D images. For 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) staining, GFP-expressing bacteria were used for biofilm formation, and immune cells were labeled with CellTracker Deep Red. After 4 days of biofilm growth, 180 μL of medium was removed from biofilms, whereupon 100 μL of 4 mM CTC (final working concentration of 1 mM) followed by 100 μL of each leukocyte population (1.5 × 106 cells) was carefully added for a final volume of 400 μL, and images were acquired within 10 min to prevent signal saturation. For 6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) staining, dsRed-expressing bacteria were used for biofilm formation, and immune cells were labeled with CellTracker Deep Red. After 4 days of biofilm growth, 180 μL of the medium was removed from biofilms, whereupon 100 μL of 40 μM CM-H2DCFDA (final working concentration of 10 μM) followed by 100 μL of each leukocyte population (1.5 × 106 cells) was carefully added for a final volume of 400 μL, and images were acquired within 40 min to prevent signal saturation.
Metabolite measurements
Supernatants were collected from biofilm following initial inoculation and daily (prior to medium replenishment) for 4 days. Supernatants were collected from planktonic cultures during initial inoculation and every 30 min up to 6 h. Enzytec UV assay kits for Liquid D-Glucose (E8140), Liquid Acetic acid (E8226), and Liquid D-/L-Lactic acid (E8240) were used for quantification.
Solutions used throughout BaSSSh-seq processing from barcoding to sequencing
PBS + RI
0.1 U/μL RI (SUPERase·In RNase Inhibitor) in 1X PBS
Tris-HCl + RI
100 mM Tris-HCl pH 8.0 (same pH used throughout), 0.1 U/μL RI
Permeabilization mix
100 mM Tris-HCl, 0.05 M EDTA, 0.25 U/μL RI, 40 μg/mL lysostaphin
2X RT mix (600 μL)
30 μL water (molecular biology grade, used throughout), 240 μL 5X RT buffer (provided with Maxima H Minus Reverse Transcriptase), 30 μL RI, 60 μL 10 mM dNTPs, 180 μL 50% PEG8000, 60 μL Maxima H Minus Reverse Transcriptase
Ligation mix (1.02 mL)
295 μL water, 250 μL 10X T4 DNA Ligase buffer (provided with T4 DNA Ligase), 75 μL T4 DNA Ligase, 25 μL RI, 375 μL 50% PEG8000
Wash buffer
0.1% Triton X-100 and 0.05 U/μL RI in 1X PBS
2X Lysis buffer
20 mM Tris-HCl, 400 mM NaCl, 100 mM EDTA, 4.4% SDS
2X BW buffer
10 mM Tris-HCl, 2 M NaCl, 1 mM EDTA, 0.1% Tween-20
S3TE-TW buffer
10 mM Tris-HCl, 0.01% Tween-20, 1 mM EDTA
S3 mix (440 μL)
111.1 μL water, 88 μL 5X RT buffer (provided with Maxima H Minus Reverse Transcriptase), 176 μL 30% PEG8000, 44 μL 10 mM dNTPs, 4.4 μL 1 mM S3_randomer, 16.5 μL Klenow Fragment (3′ → 5′ exo-)
PCR mix (440 μL)
184.4 μL water, 17.6 μL 10 μM PCR_P1, 17.6 μL 10 μM PCR_P2, 220 μL 2X KAPA HiFi HotStart ReadyMix
T.1E
10 mM Tris-HCl, 0.1 mM EDTA
Barcoding
Cell fixation, permeabilization, and counting
Fixation was achieved using 4% formaldehyde in 1X PBS overnight at 4 °C. The next morning, cells were briefly vortexed and centrifuged at 7000 × g for 5 min at 4 °C (standard centrifugation conditions used throughout the entire barcoding process) and resuspended in 1 mL cold Tris-HCl+RI. Cells were centrifuged and washed again in Tris-HCl+RI. Next, cells were resuspended in 500 μL 0.04% Tween-20 in 1X PBS and incubated on ice for 3 min. A 1 mL volume of cold PBS+RI was added before centrifuging cells and resuspending in 300 μL permeabilization mix. Cells were held in permeabilization mix at 37 °C for 15 min, with intermittent mixing. After permeabilization, 1 mL of cold PBS+RI was added before centrifugation. Cells were resuspended in 500 μL cold PBS + RI and 1 μL of 10% Tween-20 was added before another centrifugation and resuspension in 500 μL cold PBS+RI. Cells were stored on ice while counting was performed on a hemocytometer (Reichert Bright-Line, Hausser Scientific, #1492). Cells were diluted in 0.2 μm-filtered trypan blue as a contrast agent and allowed to settle for 10 min after loading onto the hemocytometer and counted using 40× magnification. A flow cytometry-based method was initially used for counting, but the background noise was too high to accurately enumerate bacterial cell numbers. Cells were diluted to ~3 × 106 per mL for barcoding. Permeabilization efficacy is shown in Supplementary Fig. 2D.
Reducing cell clumping and aggregates during barcoding
Before each barcoding step (including the first round of reverse transcription) and after ligation of the final barcode, bacterial cells were vortexed for 1 min and filtered through consecutive 10 μm and 1 μm cell strainers with gentle vacuum. Immediately prior to loading cells into the barcoding plates, cells were again vortexed for 1 min and briefly sonicated for 1–2 s.
In-cell reverse transcription (barcode 1)
Barcode 1 was added during the initial in-cell capture of RNA transcripts through random hexamer-primed reverse transcription. Wells of a 96-well plate were loaded with 6 μL of 25 μM respective round_1_barcoding oligo, 10 μL of 2X RT mix, and 4 μL of cells (at ~3 × 106 per mL after vortexing, filtering, and sonicating—Supplementary Fig. 2A–C). For experiments with multiple samples, cells from individual samples were separated for the first barcoding round for proper identification based on barcode 1 after sequencing. Several important factors influenced the selection of cell numbers for barcoding. The number of cells should not dramatically exceed the number of possible barcode combinations, or duplicated barcodes may arise in the sequencing data. However, in addition to unavoidable cell loss throughout the barcoding process, many sequenced cells (more than half) contained an inadequate number of RNA reads and were filtered out of the analysis (Supplementary Figs. 9A and 12A). This likely resulted from multiple factors including low transcriptional activity of cells in biofilm, incomplete permeabilization, and/or poor capture efficiency. The reverse transcription plate was incubated for 10 min at 23 °C and 50 min at 50 °C in a thermocycler (lid temperature set at 50 °C). Following reverse transcription, cells from all wells were collected and pooled, and 9.6 μL of 10% Triton X-100 was added prior to centrifugation. Cells were resuspended in 1 mL cold PBS+RI and vortexed, filtered, and sonicated as described above.
In-cell ligations (barcodes 2 and 3)
Barcodes 2 and 3 were added by in-cell ligation with the prior barcode. For the first ligation reaction, 2.5 μL of 30 μM respective round_2_barcoding oligo and 2.5 uL of 30 μM round2_linker oligo were pre-annealed in each well of a 96-well plate by heating to 95 °C for 2 min and cooling to 20 °C at a rate of −0.1 °C/s in a thermocycler (standard conditions for each pre-annealing step). The 1 mL of pooled cells from reverse transcription was mixed with 1.02 mL ligation mix (after vortexing, filtering, and sonicating), and 20 μL was added to each well containing the 5 μL pre-annealed barcode-linker mix. The plate was incubated at 37 °C with shaking for 30 min. Then, 5 μL of blocking mix (40 μM round2_blocking and 40 μM pre-annealed round2_blocking_HP in 2.5X T4 DNA Ligase buffer) was added to each well and incubated at 37 °C with shaking for 30 min. Cells from all wells were then pooled and vortexed, filtered, and sonicated as described above. After the final vortex and sonication, 50 μL of fresh T4 DNA Ligase was added to the cell pool. For the second ligation reaction, 3 μL of 30 μM respective round_3_barcoding oligo and 3 μL of 30 μM round3_linker oligo were pre-annealed in each well of a 96-well plate. The pooled cells from the first ligation reaction were then aliquoted at 24 μL per well. The plate was again incubated at 37 °C with shaking for 30 min. Then, 5 μL of blocking mix (45 μM round3_blocking and 45 μM pre-annealed round3_blocking_HP in 150 mM EDTA) was added to each well and incubated at 37 °C with shaking for 30 min. Cells from all wells were then pooled and vortexed, filtered, and sonicated as described above.
Final washing and cell library construction
After vortexing, filtering, and sonicating the final barcoded cell mixture, 36 μL of 10% Triton X-100 was added before centrifugation. The supernatant was removed until ~30 μL remained, to avoid aspiration of the cell pellet that was small and fragile at this step. The cell pellet was resuspended in 1.5 mL wash buffer and washing was repeated once to ensure adequate cleaning of the cells and removal of excess reagents. Next, the final cell pellet was resuspended in ~200 μL PBS+RI, and cells were counted on a hemocytometer. Aliquots of 15,000–25,000 cells in 0.5 mL tubes were then brought up to 50 μL with 1X PBS and stored at −80 °C until lysis. The number of cells per library was dependent on the experiment and how many cells were targeted for sequencing. As previously mentioned, a large portion of cells were filtered out based on low numbers of RNA reads per cell. After several sequencing runs and observing the numbers of cells filtered, cell library sizes were adjusted accordingly. Compiling cell libraries with a large number of cells is ideal for conserving reagents and costs; however, we observed that if libraries exceeded 25,000 cells, the downstream second strand synthesis step was error prone (Supplementary Fig. 4A). We believe this was caused by excess round_3_barcoding oligos that are free in solution, or not bound to captured RNA transcripts, but are still pulled out of cell lysates by their biotin tag and carried through second strand synthesis by the streptavidin beads.
Cell lysis
Cell libraries were flash thawed from −80 °C, and 50 μL 2X lysis buffer and 10 μL proteinase K solution were added. Lysis reactions were incubated at 55 °C for 2 h with shaking at 600–750 rpm. Lysates were directly stored at −80 °C until purification and second-strand synthesis.
Second strand synthesis
Purification of captured transcripts
The biotin tag on round_3_barcoding oligos facilitated transcript purification from the cell library lysates. 100 μL of Hydrophilic Streptavidin Magnetic Beads per library were aliquoted into separate 1.5 mL tubes for washing. Washing consisted of placing tubes on a magnetic stand for ~1 min until beads were sufficiently pulled out of solution, removing the liquid, and resuspending beads in ~360 μL of a particular wash buffer (note that different wash buffers were used throughout second strand synthesis). Washing was performed three times in 1X BW buffer before final resuspension of beads in 100 μL of 2X BW buffer. 2 μL of RI was added to the final suspension. Cell library lysates were briefly thawed in a 37 °C bath until the solution became clear (the solution was initially turbid and white when cool). Then, 5 μL of 10 mM PMSF was added to each lysate library and incubated at room temperature for 10 min to inhibit proteinase K activity. After 10 min, 100 μL of washed bead suspension was added to each lysate library in 0.5 mL tubes and biotin-streptavidin binding occurred during shaking at ~750 rpm at room temperature for 1 h. Beads tended to settle over time, so tubes were manually inverted every ~10 min to mix. After the 1 h binding, beads were washed in 1X BW buffer twice followed by S3TE-TW buffer twice, with 5 min shaking at ~750 rpm after resuspension during each wash.
Denaturing and second strand synthesis
Reverse transcription in the first barcoding round resulted in a hybrid RNA-cDNA construct. For second strand synthesis to occur, the captured RNA from the RNA-cDNA constructs was denatured for random priming of the cDNA (with oligo S3_randomer) to carry out second strand synthesis, as originally described32. After the final wash in S3TE-TW buffer in the previous step, beads with captured transcripts were resuspended in 500 μL 0.1 M NaOH (prepared fresh on the day of use) and incubated at room temperature for 5 min with shaking at ~750 rpm with inversion every 1 min. Beads were then washed twice in S3TE-TW buffer and twice in 10 mM Tris-HCl pH 8.0. After the last wash, beads were resuspended in 220 μL S3 mix per library and incubated at 37 °C for 1 h with shaking at 600–700 rpm, and tubes were inverted to prevent bead settling every ~10 min. After second strand synthesis, beads were washed twice in S3TE-TW buffer and once in water (molecular biology grade, used throughout).
Amplification
Amplification was required to copy the cDNA off the beads and was conducted using the PCR handle on round_3_barcoding oligos and the newly added handle from second strand synthesis. After the final water wash following second strand synthesis, beads were resuspended in 220 μL PCR mix per library. Each mix was distributed across 4–5 wells of a PCR 96-well plate and run with the following thermocycler program: 95 °C for 3 min, 3 cycles of [98 °C for 20 s, 65 °C for 45 s, 72 °C for 3 min]. The plate was then removed from the thermocycler, mixed by pipetting to resuspend settled beads, and returned to the thermocycler to repeat the same program with 2 cycles. After a total of 5 cycles, the plate was removed, briefly mixed by pipetting, and placed onto a 96-well plate magnetic stand where the beads were pulled out of solution and discarded. The remaining solution was moved into new wells for three consecutive SPRIselect bead cleanups: 0.90× with elution in 100 μL water, 0.80× with elution in 50 μL water, and 0.80× with final elution in 21 μL water. Separate wells for each library were combined back into a single well during elution in the first cleanup. Throughout cell lysis, purification, and second strand synthesis, there was a greater amount of free/unbound round_3_barcoding oligos in solution than RNA constructs. These free/unbound round_3_barcoding oligos were biotinylated, which resulted in their capture on the streptavidin magnetic beads alongside the desired RNA constructs. If left in solution for too many amplification cycles after second-strand synthesis, erroneous PCR combination events occurred leading to long polymer products(Supplementary Fig. 4A). Since the problematic round_3_barcoding oligos were shorter than RNA constructs (which become double-stranded cDNA after second strand synthesis), they could be removed based on their size by SPRIselect bead cleanups. Three consecutive cleanups were performed as described to eliminate the large excess of small products, and the elution volumes were decreased sequentially to prevent losses from using a small volume to elute from a large amount of beads. After the initial five amplification cycles and cleanup, the final 21 μL of eluted product was combined with 2 μl each of PCR_P1 and PCR_P2 primers (10 μM each) and 25 μL 2X KAPA HiFi HotStart ReadyMix and placed in the thermocycler for additional amplification with the following program: 95 °C for 3 min, 15–18 cycles of [98 °C for 20 s, 67 °C for 20 s, 72 °C for 3 min], 72 °C for 5 min. Amplification was performed until reaching a concentration of 6–10 ng/μL measured by Qubit, which was generally achieved in 20–23 total cycles (including the initial 5 cycles). After each set of cycles, a single SPRIselect bead cleanup was performed at 0.80× with elution in 21 μL water. After reaching this target concentration, fragment analysis was used to confirm a quality amplification product with fragment sizes ranging from 300 to 3000 bp as seen in the optimized quality control workflow in Supplementary Fig. 7A. Lowering the SPRIselect bead cleanup ratio reduces the number of detected cells post-sequencing, as seen in initial trials using 0.60× ratios in second strand synthesis and the subsequent rRNA depletion steps (Supplementary Fig. 4B–D).
Note on library concentrations and pooling
A target library concentration of 6–10 ng/μL was sufficient for downstream processing with reserve in case there was a need to return to the previous step. Moving through each step following amplification, at least 25% of the library volume was carried forward to ensure representation of full library complexity. After second strand synthesis, the libraries can be safely pooled (if desired) since the issues described in Supplementary Fig. 4A have been circumvented. Pooling helps to reduce costs and increase cell numbers for analysis when implementing a stringent reads per cell cutoff, especially for biofilm cells that are less metabolically and transcriptionally active, where many are filtered out of the analysis (as illustrated in Supplementary Figs. 9A and 12A). Pooled cells can be separately dual-indexed for multiplexed libraries to keep samples separated and reduce the probability of barcode duplication. If pooling, the 6–10 ng/µL concentrations for individual cell libraries remains a good target, and at least 25% of each individual sample should be combined for the pooled mix.
rRNA depletion: dual-strand subtractive hybridization
Forward and reverse oligo master mixes
Sets of biotinylated oligos designed against 23S, 16S, and 5S fragments for rRNA depletion from double-stranded cDNA (rRNA_dep_Fwd and rRNA_dep_Rev) were each suspended at 100 μM in water from the solid desalted form. An equal volume (5 μL) of all 21 oligos from forward or reverse sets was pooled into a single master mix for either forward (MM-Fwd) or reverse (MM-Rev) strand depletion, being careful not to combine forward and reverse oligos. In the pooled mixes with equal volume of 21 forward or reverse oligos, the concentration of each oligo was 4.76 μM.
Forward and reverse depletion mixes
Working mixes for depletion of forward and reverse rRNA fragments (dep-M-Fwd and dep-M-Rev) were comprised of SSC buffer (20X), EDTA (100 mM), T.1E, and MM-Fwd or MM-Rev. The specific amounts of each component was calculated using the Excel calculator tool developed by Culviner et al. based on total RNA input, desired oligo:RNA and bead:oligo ratios, and total reaction volume33. In our implementation of dual-strand subtractive hybridization, total RNA input was variable based on the amplified library concentration following second strand synthesis (generally in the range of 200–250 ng), oligo:RNA ratio was set at 5, bead:oligo ratio was set at 18, and the reaction volume was set at 70 μL. In the calculator tool, the number of oligos should be set at 21 and the bead capacity should be set to 1.60 × 10−12 mol/μL if using NEB Hydrophilic Streptavidin Magnetic Beads. Results from initial trials with varying oligo:RNA and bead:oligo ratios can be found in Supplementary Fig. 5. Larger ratios lead to greater rRNA depletion but use significantly more oligo and bead reagents. We selected our values by considering both depletion efficiency and cost.
Example—An amplified library after second strand synthesis is 29 µL at a concentration of 8.28 ng/μL. If nearly the full volume is used, then the total RNA input can be set at 230 ng (requiring 27.8 μL of the library), oligo:RNA = 5, bead:oligo = 18, and reaction volume at 70 μL. The forward depletion mix (dep-M-Fwd) is made according to the calculator tool33 as 43.75 μL 20X SSC, 8.75 μL 100 mM EDTA, 3.67 μL MM-Fwd, and 124.4 μL T.1E. Note that the total volume of dep-M-Fwd is about 25× larger than what is required; however, this large volume is used such that the volume of MM-Fwd is accurately measurable. To calculate the requirements for the reverse depletion mix (dep-M-Rev), the theoretical concentration in 25 μL is input as the concentration (230 ng/25 μL = 9.2 ng/μL) with all other settings held uniform. This is because depletion of the reverse strand is initiated with 25 μL volume. The dep-M-Rev mix is made as 43.75 μL 20X SSC, 8.75 μL 100 mM EDTA, 3.67 μL MM-Rev, and 193.83 μL T.1E. Again, the total volume of dep-M-Rev is about 25× larger than what is required for measurement accuracy.
Prepare streptavidin beads
The volume of streptavidin beads was also determined by the calculator tool. Since our method depleted from both the forward and reverse strands, the amount of beads was doubled.
Example (continued from above)—331 μL of beads are used. The beads are washed three times with ~500 μL of 5X SSC, then resuspended in 70 μL 5X SSC and kept at room temperature until used.
Forward strand depletion and cleanup
The library was first denatured in a thermocycler by heating to 95 °C for 2 min and then cooling to 20 °C. A 96-well PCR plate was used for all depletion steps. Then, the dep-M-Fwd mix was added as directed in the calculator tool to achieve a final volume of 35 μL. Annealing of the depletion oligos to cDNA template was carried out in a thermocycler with the following program: 95 °C for 3 min, 98 °C for 20 s, 70 °C for 5 min, decrease to 20 °C at a rate of −0.1 °C/s. Following annealing, the rRNA constructs were depleted by adding 35 μL washed streptavidin beads and mixing by pipetting at least 25×. The bead mixture was incubated at room temperature with shaking (~500–700 rpm) and intermittent mixing with a pipette for 10 min. After a final pipette mixing step, bead mixtures were incubated in a thermocycler at 50 °C for 5 min (lid temperature reduced to 50 °C). Mixtures were then immediately set on a magnetic plate and the supernatants transferred to new wells after the beads had adequately settled (beads were discarded). SPRIselect bead cleanup was then performed on the supernatants at 0.90× with elution in 25 μL water.
Example (continued from above)—27.8 μL of library is denatured, then 7.2 μL of dep-M-Fwd is added and run on the annealing program.
Reverse strand depletion and cleanup
The 25 μL volume from forward strand depletion and cleanup was heated to 95 °C for 2 min and then cooled to 20 °C. Then, 10 μL dep-M-Rev mix was added, and annealing of the depletion oligos to cDNA template was performed in a thermocycler with the same annealing program described for forward strand depletion. As for the forward strand, 35 μL of washed streptavidin beads were added and incubated for 10 min at room temperature with mixing before heating at 50 °C for 5 min and removing the beads via magnet. A 0.90× SPRIselect bead cleanup was then performed with elution in 21 μL water.
Amplification
The final eluted 21 μL of rRNA-depleted product was combined with 2 μl each of PCR_P1 and PCR_P2 primers (10 μM each) and 25 μL 2X KAPA HiFi HotStart ReadyMix and amplified with the following thermocycler program: 95 °C for 3 min, 4–5 cycles of [98 °C for 20 s, 67 °C for 20 s, 72 °C for 3 min], 72 °C for 5 min. Amplification was performed until reaching a concentration of 10–20 ng/μL measured by Qubit, which was generally achieved in 4–5 total cycles. A 0.90× SPRIselect bead cleanup was then performed with elution in 21 μL water. Fragment analysis was used to confirm a quality amplification product with fragment sizes ranging from 300–3000 bp (Supplementary Fig. 7A). In comparison to the size distributions following second strand synthesis, size distributions after rRNA depletion were shifted towards the 700–3000 bp range. As an additional quality check, qPCR was performed side-by-side on samples before and after depletion. Analysis of 23S and 16S rRNA abundance in relation to a control gene (gyrB) showed substantial rRNA depletion with minimal alterations to the control gene (Supplementary Fig. 7B). The qPCR was performed with Luna Universal qPCR Master Mix from NEB and primer sets for gyrB (gyrB_Fwd and gyrB_Rev), 23S rRNA (23S_Fwd and 23S_Rev), and 16S rRNA (16S_Fwd and 16S_Rev).
Library prep and sequencing
Fragmentation and cleanup
Reagents used in fragmentation were from the sparQ DNA fragment and library prep kit (Quantabio). A mix of 5X DNA Frag & Polishing Enzyme Mix (10 μL per library) and 10X DNA Frag & Polishing Buffer (5 μL per library) was first prepared and kept on ice. Next, 15 μL of this mix was added to 150–250 ng rRNA-depleted sample library in 35 μL water, for a total volume of 50 μL which was also held on ice. The 50 μL fragmentation mix was quickly transferred to a pre-chilled thermocycler and run on the following program: 4 °C for 1 min, 32 °C for 8 min (fragmentation step), 65 °C for 30 min, and 4 °C hold. The samples were immediately removed and stored on ice before quickly initiating a double-side 0.825–0.45× SPRIselect bead cleanup. Final product was eluted in 30 μL water, then brought up to 50.5 μL with water. Fragmentation temperature and time were explored in initial trials as shown in Supplementary Fig. 6A, and an overview of library size distributions from input to fragmentation product, and following the double-sided SPRIselect bead cleanup is provided in Supplementary Fig. 6B for guidance.
Ligation
Reagents used in ligation were also from the sparQ DNA fragment and library prep kit (Quantabio), with additional custom oligos. A mix of 5X DNA Rapid Ligation Buffer (20 μL per library), DNA Ligase (10 μL per library), and water (17.5 μL per library) was prepared and kept on ice. The adapter duplex consisting of adapter_duplex_Top and adapter_duplex_Bott was pre-annealed by heating an adapter mix (100 μM each in 50 μM NaCl) to 95 °C and cooling to 20 °C at a rate of −0.1 °C/s. Then, 2 μL of the pre-annealed adapter duplex mix was added to each 50.5 μL library (after double-sided bead cleanup) on ice. Finally, 47.5 μL of the ligation mix was added to each 52.5 μL library with adapters, and the 100 μL solution was added to a thermocycler for incubation at 20 °C for 15 min (with unheated lid). The ligation reactions were immediately removed from the thermocycler and SPRIselect cleaned two consecutive times at 0.81×, first eluting in 50 μL water and then 21 μL water on the final cleanup.
Amplification
The 21 μL cleaned ligation product was combined with 2 μL each of i5 primer (seq_i501/2) and i7 primer (seq_i701/2) (10 μM each) and 25 μL 2X KAPA HiFi HotStart ReadyMix and amplified with the following thermocycler program: 95 °C for 3 min, 4–6 cycles of [98 °C for 20 s, 67 °C for 20 s, 72 °C for 3 min], 72 °C for 5 min. The number of cycles depended on the input amount, where 250 ng input required only 4 cycles, whereas 150 ng input required 6 cycles. Next, two consecutive double-sided SPRIselect bead cleanups were performed, first at 0.75–0.45× and then at 0.775–0.425×. Two cleanups were necessary to adequately remove fragments that were too small or large. Fragment analysis was performed for final library quality control as seen in the optimized quality control workflow in Supplementary Fig. 7A. The input quantity, fragmentation time, and number of cycles all affect the resulting sequencing library concentration. A representative selection of metrics and how they relate to final sequencing library concentration is provided in Supplementary Fig. 6C. Ideally, the concentration should be kept near the minimum amount required to load the sequencer to ensure full library diversity is represented, as seen in example runs 7 and 8 in Supplementary Fig. 6C.
Library quantification
NEBNext Library Quant Kit for Illumina was used for qPCR-based library quantification (Supplementary Fig. 7C). With the described library prep protocol, final libraries were in the range of 1–2 nM. We found it beneficial to keep library concentrations low to maximize the amount of each library loaded on the sequencer, which ensured capturing full library diversity.
Sequencing
The datasets presented were sequenced on an Illumina NextSeq 550 series sequencer with v2.5 150-cycle mid-output kit. Read 1 (insert/transcript) was allocated 63 cycles, and read 2 (barcodes) was allocated 89 cycles, 3 extra bases than the full-length barcodes that are 86 bp.
Count matrix generation
Remove adapters and quality filter
Cutadapt177 was used to identify and remove sequencing adapters from the ends of read 1 and read 2 (AGATCGGAAGAGCACACGTCTGAACTCC and ACTGTCTCTTATACACATCT, respectively) and filter out reads with quality scores <10.
Extract UMIs
UMI sequences were extracted from read 2 with UMI-tools178 using the extract command.
Demultiplexing
Cutadapt was used to demultiplex reads by the 5′-end into unique combinations of three barcodes beginning with barcode 3, then barcode 2, and barcode 1. A python script was used to loop Cutadapt through all files, and each unique barcode combination was output into a separate file. Barcode 3 was demultiplexed as the 8-nucleotide barcode and linker sequence between barcodes 2 and 3 (totaling 38 nucleotides) and processed as a non-internal 5′ adapter with minimum overlap of 35 and error tolerance set at 0.15. Barcode 2 was demultiplexed as the 8-nucleotide barcode and linker sequence between barcodes 1 and 2 (totaling 30 nucleotides) and processed as a non-internal 5′ adapter with minimum overlap of 25 and error tolerance set at 0.2. Barcode 1 was demultiplexed as the 8-nucleotide barcode and processed as a non-internal 5′ adapter with minimum overlap of 6. After demultiplexing, cells were separated by sample based on barcode 1.
Alignment
After demultiplexing, only read 1 sequences were maintained for processing. Reads were mapped to the annotated S. aureus USA300 FPR3757 genome (NCBI)179 using STAR180. As previously reported, we ignored splicing detection and retained multimapping reads, as bacterial genomes are known to contain overlapping coding sequences21. Additionally, many reads had either full or partial fragments of the second strand synthesis oligo (S3_randomer, 34 nucleotides), which reduced the portion of the read available for mapping. Therefore, the options -outFilterMatchNminOverLread and -outFilterScoreMinOverLread were reduced to 0.2 to increase tolerance for smaller fractions of the aligning read. This adjustment was necessary to prevent loss of transcriptional information and data bias; however, it also led to inherent background noise as extraneous multimapping reads were inevitably identified.
Counting
The featureCounts181 package of Subread was used to annotate and enumerate transcript features after alignment. Multimapping reads were assigned a fractional count, which was necessary for accurately quantifying rRNA reads that aligned to multiple regions in the genome, as well as to attenuate background noise from spurious alignments.
Collapse UMIs
The dedup command within UMI-tools was used to deduplicate reads based on UMI and mapping coordinates.
Count matrix
A matrix of cells-by-genes was filled with gene counts per cell, for input into Scanpy.
Data processing and clustering
Preprocessing, filtering, and normalization
Data analysis was performed in Scanpy182. All rRNA genes were removed, and cells were filtered based on total mRNA and tRNA (non-rRNA) expression levels. For the biofilm vs. planktonic comparison experiment, biofilm cells having ≥7 and planktonic cells having ≥28 non-rRNA reads per cell were maintained for analysis, totaling 3680 and 4231 cells respectively (Supplementary Figs. 9A and 10A). Resulting rRNA, mRNA, and tRNA metrics are shown in Supplementary Fig. 9B–C. For the biofilm-leukocyte co-culture experiment, cells from all conditions were kept for analysis if they contained ≥15 non-rRNA reads per cell, totaling 4655, 4544, 4780, and 6125 cells for the biofilm control, biofilm + MΦs, biofilm + G-MDSCs, and biofilm + PMNs respectively (Supplementary Fig. 12A). Resulting rRNA, mRNA, and tRNA metrics are shown in Supplementary Fig. 12B–C. All cells were uniformly normalized to 104 total counts and log+1 transformed.
Clustering
For the biofilm and planktonic comparison experiment, cells from each growth state were combined and principal components calculated from highly variable genes (min_mean = 0.00625 and min_disp = 0.25). Nearest neighbors were determined and integrated with BBKNN alignment (neighbors_within_batch = 9 and n_pcs = 4)64. Neighbors were UMAP embedded (min_dist = 0.24 and spread = 0.21) and clustered with the Leiden algorithm (resolution = 0.205). For the biofilm-leukocyte co-culture experiment, cells from each condition were first independently analyzed with principal components calculated from highly variable genes (min_mean = 0.00625 and min_disp = 0.25). Nearest neighbors were determined (n_neighbors = 12 and n_pcs = 7), UMAP embedded (min_dist = 0.5 and spread = 1), and clustered with the Leiden algorithm (resolution = 0.15). For comparison of biofilm responses to MΦs, G-MDSCs, and PMNs, biofilm cells from each condition were projected onto the UMAP of the biofilm control using the ingest function of Scanpy. The noted clustering settings were chosen to optimize resolved sets of marker genes, which were used as a quality control readout (Supplementary Fig. 9D and Supplementary Fig. 13).
Differential expression
The MAST algorithm65 was integrated with the Scanpy workflow through the rpy2, anndata2ri, and sc_toolbox packages and IPython interface in Jupyter notebook183. MAST was used for all differential expression analyses. A known caveat of MAST is that log2 fold-change values can be small; therefore, marginal differences cannot be disregarded as insignificant.
iModulon and transcriptional regulatory network analyses
Gene sets were downloaded from the S. aureus Precise165 dataset on the iModulonDB database39 as determined in ref. 37, and USA300_TCH1516 annotations were converted to USA300_FPR3757 orthologues using AureoWiki databases184. On a per cell basis, raw gene expression was summed for all genes within each iModulon, forming an expression score. Next, raw iModulon scores were summed for all iModulons comprising a transcriptional regulatory network category, forming a regulatory category score. The iModulon and transcriptional regulatory network scores were uniformly normalized to 104 total counts and log+1 transformed separately from the individual genes.
Trajectory analysis
The Palantir package was used for trajectory analysis40. Diffusion maps (n_components = 5) were first determined, and MAGIC imputation101 (from within the Palantir package) was used for resolving expression trends. Imputed expression was then used for all UMAP visualizations. Trajectory analysis (num_waypoints = 500) was then performed with an indicated starting cell and terminal states, and importantly altering the cell of origin did not affect trajectory outcome. Detailed initiating and terminal state selections are shown in Supplementary Fig. 11. Pearson correlations were calculated for the imputed expression of each gene over the Palantir-calculated pseudotime variable. Top lists of positively and negatively correlated genes with pseudotime are shown in Supplementary Data 8–9.
Statistical analyses
The statistical tests for all BaSSSh-seq experiments were performed in python 3 (3.10.12) within Jupyter notebook or GraphPad Prism (10.0.2). Statistical details can be found in figures and legends where appropriate.
Reporting summary
Further information on research design is available in Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke F32NS126302 to L.E.K. and the National Institute of Allergy and Infectious Diseases 3P01AI083211 (Project 4) to T.K. The UNMC Genomics Core Facility receives partial support from the National Institute for General Medical Sciences INBRE P20GM103427-19. The UNMC Flow Cytometry Research Facility is administrated through the Office of the Vice Chancellor for Research and supported by state funds from the Nebraska Research Initiative. Both the Genomics Core and Flow Cytometry Research Facilities are partially supported by The Fred & Pamela Buffett Cancer Center’s National Cancer Institute Cancer Support Grant P30CA036727. We thank Drs. Paul D. Fey and Vinai C. Thomas for their expert discussions and suggestions on data analysis and interpretation. We thank Jennifer Endres for support with equipment, reagents, and database curation. We acknowledge Dr. Eric Tom and Nichole Brandquist for their technical assistance in sequencing library QC and confocal imaging, respectively. We acknowledge Drs. Anna Kuchina and Sydney Blattman for sharing their expertise during the initial experimental setup. We appreciate the discussions and suggestions from current and former members of the Kielian Laboratory and others at UNMC including Drs. Blake Bertrand, Christopher Horn, Gunjan Kak, and Prabhakar Arumugam, as well as Zachary Van Roy and Cleofes Sarmiento.
Author contributions
L.E.K. and T.K. designed the study. L.E.K. conducted experiments, performed data analysis, and wrote the manuscript. Both authors edited and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Sequencing data and processed count matrices have been deposited in the GEO database under accession code GSE270986. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lee E. Korshoj, Email: lee.korshoj@unmc.edu
Tammy Kielian, Email: tkielian@unmc.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54581-8.
References
- 1.Nelson, R. E. et al. National Estimates of Healthcare Costs associated with multidrug-resistant bacterial infections among hospitalized patients in the United States. Clin. Infect. Dis.72, S17–S26 (2021). [DOI] [PubMed] [Google Scholar]
- 2.O’Connell, K. M. et al. Combating multidrug-resistant bacteria: current strategies for the discovery of novel antibacterials. Angew. Chem. Int. Ed. Engl.52, 10706–10733 (2013). [DOI] [PubMed] [Google Scholar]
- 3.van Duin, D. & Paterson, D. L. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect. Dis. Clin. North Am.30, 377–390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naghavi, M. et al. Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet404, 1199–1226 (2024). [DOI] [PubMed]
- 5.Turner, N. A. et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol.17, 203–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong, N. W. M., van Kessel, K. P. M. & van Strijp, J. A. G. Immune evasion by Staphylococcus aureus. Microbiol. Spectr.7. 10.1128/microbiolspec.GPP3-0061-2019 (2019). [DOI] [PubMed]
- 7.Schilcher, K. & Horswill, A. R. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev.84, e00026–00019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vestby, L. K., Gronseth, T., Simm, R. & Nesse, L. L. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics9, 59 (2020). [DOI] [PMC free article] [PubMed]
- 9.Li, P., Yin, R., Cheng, J. & Lin, J. Bacterial biofilm formation on biomaterials and approaches to its treatment and prevention. Int. J. Mol. Sci.24, 11680 (2023). [DOI] [PMC free article] [PubMed]
- 10.Jovic, D. et al. Single-cell RNA sequencing technologies and applications: a brief overview. Clin. Transl. Med.12, e694 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldrich, A. L., Horn, C. M., Heim, C. E., Korshoj, L. E. & Kielian, T. Transcriptional diversity and niche-specific distribution of leukocyte populations during Staphylococcus aureus craniotomy-associated biofilm infection. J. Immunol.206, 751–765 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bossel Ben-Moshe, N. et al. Predicting bacterial infection outcomes using single cell RNA-sequencing analysis of human immune cells. Nat. Commun.10, 3266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menousek, J. et al. Transcriptional profiling of phagocytic leukocytes and microglia reveals a critical role for reactive oxygen species in biofilm containment during Staphylococcus aureus craniotomy infection. J. Immunol.209, 1973–1986 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papalexi, E. & Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol.18, 35–45 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Reyes, M. et al. An immune-cell signature of bacterial sepsis. Nat. Med.26, 333–340 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan, M. A. & Rosenthal, A. Z. Single-cell RNA sequencing elucidates the structure and organization of microbial communities. Front. Microbiol.12, 713128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homberger, C., Barquist, L. & Vogel, J. Ushering in a new era of single-cell transcriptomics in bacteria. Microlife3, uqac020 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imdahl, F. & Saliba, A. E. Advances and challenges in single-cell RNA-seq of microbial communities. Curr. Opin. Microbiol57, 102–110 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Blattman, S. B., Jiang, W., Oikonomou, P. & Tavazoie, S. Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nat. Microbiol.5, 1192–1201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homberger, C., Hayward, R., Barquist, L. & Vogel, J. Improved bacterial single-cell RNA-seq through automated MATQ-seq and Cas9-based removal of rRNA reads. mBio14, e0355722 (2023). [DOI] [PMC free article] [PubMed]
- 21.Kuchina, A. et al. Microbial single-cell RNA sequencing by split-pool barcoding. Science371, eaba5257 (2021). [DOI] [PMC free article] [PubMed]
- 22.Ma, P. et al. Bacterial droplet-based single-cell RNA-seq reveals antibiotic-associated heterogeneous cellular states. Cell186, 877–891 e814 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNulty, R. et al. Probe-based bacterial single-cell RNA sequencing predicts toxin regulation. Nat. Microbiol.8, 934–945 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pountain, A. W. et al. Transcription-replication interactions reveal bacterial genome regulation. Nature626, 661–669 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, Z. et al. Droplet-based high-throughput single microbe RNA sequencing by smRandom-seq. Nat. Commun.14, 5130 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, B. et al. Single-cell massively-parallel multiplexed microbial sequencing (M3-seq) identifies rare bacterial populations and profiles phage infection. Nat. Microbiol.8, 1846–1862 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imdahl, F., Vafadarnejad, E., Homberger, C., Saliba, A. E. & Vogel, J. Single-cell RNA-sequencing reports growth-condition-specific global transcriptomes of individual bacteria. Nat. Microbiol.5, 1202–1206 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivatsan, S. R. et al. Massively multiplex chemical transcriptomics at single-cell resolution. Science367, 45–51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes, T. K. et al. Second-strand synthesis-based massively parallel scRNA-seq reveals cellular states and molecular features of human inflammatory skin pathologies. Immunity53, 878–894.e877 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Culviner, P. H., Guegler, C. K. & Laub, M. T. A simple, cost-effective, and robust method for rRNA depletion in RNA-sequencing studies. mBio11, e00010–e00020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Morais, S. D., Kak, G., Menousek, J. P. & Kielian, T. Immunopathogenesis of craniotomy infection and niche-specific immune responses to biofilm. Front. Immunol.12, 625467 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heim, C. E. et al. Human prosthetic joint infections are associated with myeloid-derived suppressor cells (MDSCs): implications for infection persistence. J. Orthop. Res.36, 1605–1613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horn, C. M. & Kielian, T. Crosstalk between staphylococcus aureus and innate immunity: focus on immunometabolism. Front. Immunol.11, 621750 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poudel, S. et al. Coordination of CcpA and CodY regulators in staphylococcus aureus USA300 strains. mSystems7, e0048022 (2022). [DOI] [PMC free article] [PubMed]
- 38.Poudel, S. et al. Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators, and role in key physiological response. Proc. Natl. Acad. Sci. USA117, 17228–17239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rychel, K. et al. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Res.49, D112–D120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol.37, 451–460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc.9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Kapteyn, J., He, R., McDowell, E. T. & Gang, D. R. Incorporation of non-natural nucleotides into template-switching oligonucleotides reduces background and improves cDNA synthesis from very small RNA samples. BMC Genom.11, 413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertrand, B. P. et al. Role of Staphylococcus aureus formate metabolism during prosthetic joint infection. Infect. Immun.90, e0042822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juillot, D. et al. A high-content microscopy screening identifies new genes involved in cell width control in Bacillus subtilis. mSystems6, e0101721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, S., Wu, L. J. & Errington, J. Microfluidic time-lapse analysis and reevaluation of the Bacillus subtilis cell cycle. Microbiologyopen.8, e876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng, A. G., Missiakas, D. & Schneewind, O. The giant protein Ebh is a determinant of Staphylococcus aureus cell size and complement resistance. J. Bacteriol.196, 971–981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heimberg, G., Bhatnagar, R., El-Samad, H. & Thomson, M. Low dimensionality in gene expression data enables the accurate extraction of transcriptional programs from shallow sequencing. Cell Syst.2, 239–250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollen, A. A. et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol.32, 1053–1058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, A. R. et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods11, 41–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X., Xu, C. & Yosef, N. Simulating multiple faceted variability in single cell RNA sequencing. Nat. Commun.10, 2611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Betancur, J. C. & Lopez, D. Cell heterogeneity in Staphylococcal communities. J. Mol. Biol.431, 4699–4711 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Hart, J. W., Waigh, T. A., Lu, J. R. & Roberts, I. S. Microrheology and spatial heterogeneity of Staphylococcus aureus biofilms modulated by hydrodynamic shear and biofilm-degrading enzymes. Langmuir35, 3553–3561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moormeier, D. E. & Bayles, K. W. Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol.104, 365–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rani, S. A. et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J. Bacteriol.189, 4223–4233 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart, P. S. & Franklin, M. J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol.6, 199–210 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Moormeier, D. E., Bose, J. L., Horswill, A. R. & Bayles, K. W. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio5, e01341–01314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moormeier, D. E. et al. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl. Environ. Microbiol.79, 3413–3424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadykov, M. R. & Bayles, K. W. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr. Opin. Microbiol.15, 211–215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker, P., Hufnagle, W., Peters, G. & Herrmann, M. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol.67, 2958–2965 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kot, B., Sytykiewicz, H. & Sprawka, I. Expression of the biofilm-associated genes in methicillin-resistant Staphylococcus aureus in biofilm and planktonic conditions. Int. J. Mol. Sci.19, 3487 (2018). [DOI] [PMC free article] [PubMed]
- 61.Resch, A. et al. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics6, 1867–1877 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Resch, A., Rosenstein, R., Nerz, C. & Gotz, F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol.71, 2663–2676 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol.21, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polanski, K. et al. BBKNN: fast batch alignment of single-cell transcriptomes. Bioinformatics36, 964–965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol.16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abushahba, M. F., Mohammad, H. & Seleem, M. N. Targeting multidrug-resistant Staphylococci with an anti-rpoA peptide nucleic acid conjugated to the HIV-1 TAT cell penetrating peptide. Mol. Ther. Nucleic Acids5, e339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Besier, S., Ludwig, A., Brade, V. & Wichelhaus, T. A. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol. Microbiol.47, 463–469 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Widjaja, M. et al. Elongation factor Tu is a multifunctional and processed moonlighting protein. Sci. Rep.7, 11227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bosch, M. E. et al. Staphylococcus aureus ATP synthase promotes biofilm persistence by influencing innate immunity. mBio11, e01581-20 (2020). [DOI] [PMC free article] [PubMed]
- 70.Hammer, N. D., Schurig-Briccio, L. A., Gerdes, S. Y., Gennis, R. B. & Skaar, E. P. CtaM Is Required for Menaquinol Oxidase aa3 Function in Staphylococcus aureus. mBio7, e00823-16 (2016). [DOI] [PMC free article] [PubMed]
- 71.Bowman, L., Zeden, M. S., Schuster, C. F., Kaever, V. & Grundling, A. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J. Biol. Chem.291, 26970–26986 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang, Y. H., Guan, H. H., Chen, C. J. & Huang, C. Y. Staphylococcus aureus single-stranded DNA-binding protein SsbA can bind but cannot stimulate PriA helicase. PLoS ONE12, e0182060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oun, S. et al. The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol.10, 157–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roux, A., Todd, D. A., Velazquez, J. V., Cech, N. B. & Sonenshein, A. L. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J. Bacteriol.196, 1184–1196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van den Berg, S. et al. A human monoclonal antibody targeting the conserved staphylococcal antigen IsaA protects mice against Staphylococcus aureus bacteremia. Int J. Med. Microbiol.305, 55–64 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Gaupp, R., Ledala, N. & Somerville, G. A. Staphylococcal response to oxidative stress. Front. Cell Infect. Microbiol.2, 33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kehl-Fie, T. E. et al. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect. Immun.81, 3395–3405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schultz, B. J., Snow, E. D. & Walker, S. Mechanism of D-alanine transfer to teichoic acids shows how bacteria acylate cell envelope polymers. Nat. Microbiol.8, 1318–1329 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Resch, A., Fehrenbacher, B., Eisele, K., Schaller, M. & Gotz, F. Phage release from biofilm and planktonic Staphylococcus aureus cells. FEMS Microbiol. Lett.252, 89–96 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Chang, T. L. et al. Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling circle replication. J. Biol. Chem.277, 45880–45886 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Cuny, C., Layer, F., Kock, R., Werner, G. & Witte, W. Methicillin susceptible Staphylococcus aureus (MSSA) of clonal complex CC398, t571 from infections in humans are still rare in Germany. PLoS ONE8, e83165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deghorain, M. & Van Melderen, L. The Staphylococci phages family: an overview. Viruses4, 3316–3335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ha, K. P. & Edwards, A. M. DNA repair in Staphylococcus aureus. Microbiol Mol. Biol. Rev.85, e0009121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iyer, L. M., Koonin, E. V. & Aravind, L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genom.3, 8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reslane, I. et al. Glutamate-dependent arginine biosynthesis requires the inactivation of spoVG, sarA, and ahrC in Staphylococcus aureus. J. Bacteriol.206, e0033723 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Girish, T. S., Sharma, E. & Gopal, B. Structural and functional characterization of Staphylococcus aureus dihydrodipicolinate synthase. FEBS Lett.582, 2923–2930 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Lensmire, J. M. et al. The Staphylococcus aureus cystine transporters TcyABC and TcyP facilitate nutrient sulfur acquisition during infection. Infect. Immun.88, e00690-19 (2020). [DOI] [PMC free article] [PubMed]
- 88.Soutourina, O. et al. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol.73, 194–211 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Wang, X. et al. Involvement of small colony variant-related heme biosynthesis genes in Staphylococcus aureus persister formation in vitro. Front. Microbiol.12, 756809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hiron, A., Borezee-Durant, E., Piard, J. C. & Juillard, V. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol.189, 5119–5129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shibamura-Fujiogi, M. et al. GltS regulates biofilm formation in methicillin-resistant Staphylococcus aureus. Commun. Biol.5, 1284 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Satiaputra, J., Shearwin, K. E., Booker, G. W. & Polyak, S. W. Mechanisms of biotin-regulated gene expression in microbes. Synth. Syst. Biotechnol.1, 17–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sitthisak, S., Knutsson, L., Webb, J. W. & Jayaswal, R. K. Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology153, 4274–4283 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Marincola, G. et al. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol. Microbiol.85, 817–832 (2012). [DOI] [PubMed] [Google Scholar]
- 95.Abraham, N. M. & Jefferson, K. K. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology158, 1504–1512 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gries, C. M., Biddle, T., Bose, J. L., Kielian, T. & Lo, D. D. Staphylococcus aureus fibronectin binding protein a mediates biofilm development and infection. Infect. Immun.88, e00859-19 (2020). [DOI] [PMC free article] [PubMed]
- 97.Lacey, K. A., Mulcahy, M. E., Towell, A. M., Geoghegan, J. A. & McLoughlin, R. M. Clumping factor B is an important virulence factor during Staphylococcus aureus skin infection and a promising vaccine target. PLoS Pathog.15, e1007713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin, M. H., Shu, J. C., Huang, H. Y. & Cheng, Y. C. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS ONE7, e34388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muthukrishnan, G. et al. Exoproteome of Staphylococcus aureus reveals putative determinants of nasal carriage. J. Proteome Res.10, 2064–2078 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao, X. et al. Exoproteome Heterogeneity among Closely Related Staphylococcus aureus t437 Isolates and Possible Implications for Virulence. J. Proteome Res.18, 2859–2874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell174, 716–729.e727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rudra, P. & Boyd, J. M. Metabolic control of virulence factor production in Staphylococcus aureus. Curr. Opin. Microbiol.55, 81–87 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fuller, J. R. et al. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell Infect. Microbiol.1, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Richardson, A. R., Libby, S. J. & Fang, F. C. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science319, 1672–1676 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Spahich, N. A., Vitko, N. P., Thurlow, L. R., Temple, B. & Richardson, A. R. Staphylococcus aureus lactate- and malate-quinone oxidoreductases contribute to nitric oxide resistance and virulence. Mol. Microbiol.100, 759–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hammer, N. D. et al. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio4, e00241-13 (2013). [DOI] [PMC free article] [PubMed]
- 107.Leibig, M. et al. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J. Bacteriol.193, 952–962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Falugi, F., Kim, H. K., Missiakas, D. M. & Schneewind, O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio4, e00575–00513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Crosby, H. A. et al. The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog.12, e1005604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kukita, K. et al. Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect. Immun.81, 1870–1879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang, Y. et al. Benchmarking algorithms for pathway activity transformation of single-cell RNA-seq data. Comput. Struct. Biotechnol. J.18, 2953–2961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gene Ontology, C. The gene ontology project in 2008. Nucleic Acids Res.36, D440–D444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thomas, P. D. et al. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci.31, 8–22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Houston, P., Rowe, S. E., Pozzi, C., Waters, E. M. & O’Gara, J. P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun.79, 1153–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Periasamy, S. et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA109, 1281–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cao, Z., Casabona, M. G., Kneuper, H., Chalmers, J. D. & Palmer, T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol.2, 16183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seif, Y. et al. A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types. PLoS Comput. Biol.15, e1006644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cui, L. et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother.54, 5222–5233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith, J. L. & Grossman, A. D. In vitro whole genome DNA binding analysis of the bacterial replication initiator and transcription factor DnaA. PLoS Genet.11, e1005258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Troitzsch, A. et al. Carbon source-dependent reprogramming of anaerobic metabolism in Staphylococcus aureus. J. Bacteriol.203, e00639-20 (2021). [DOI] [PMC free article] [PubMed]
- 121.Heim, C. E., West, S. C., Ali, H. & Kielian, T. Heterogeneity of Ly6G(+) Ly6C(+) Myeloid-Derived Suppressor Cell Infiltrates during Staphylococcus aureus Biofilm Infection. Infect. Immun.86, e00684-18 (2018). [DOI] [PMC free article] [PubMed]
- 122.Van Roy, Z., Shi, W., Kak, G., Duan, B. & Kielian, T. Epigenetic regulation of leukocyte inflammatory mediator production dictates Staphylococcus aureus craniotomy infection outcome. J. Immunol.211, 414–428 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yamada, K. J. & Kielian, T. Biofilm-leukocyte cross-talk: impact on immune polarization and immunometabolism. J. Innate Immun.11, 280–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bernthal, N. M. et al. A mouse model of post-arthroplasty Staphylococcus aureus joint infection to evaluate in vivo the efficacy of antimicrobial implant coatings. PLoS ONE5, e12580 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Niska, J. A. et al. Vancomycin-rifampin combination therapy has enhanced efficacy against an experimental Staphylococcus aureus prosthetic joint infection. Antimicrob. Agents Chemother.57, 5080–5086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Spitzfaden, C. et al. The structure of ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J. Mol. Biol.295, 105–115 (2000). [DOI] [PubMed] [Google Scholar]
- 127.Hartmann, T. et al. The catabolite control protein E (CcpE) affects virulence determinant production and pathogenesis of Staphylococcus aureus. J. Biol. Chem.289, 29701–29711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Downer, R., Roche, F., Park, P. W., Mecham, R. P. & Foster, T. J. The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem.277, 243–250 (2002). [DOI] [PubMed] [Google Scholar]
- 129.Grundling, A. & Schneewind, O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA104, 8478–8483 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Visai, L. et al. Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology155, 667–679 (2009). [DOI] [PubMed] [Google Scholar]
- 131.Parraga Solorzano, P. K., Bastille, T. S., Radin, J. N. & Kehl-Fie, T. E. A manganese-independent aldolase enables staphylococcus aureus to resist host-imposed metal starvation. mBio14, e0322322 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Radin, J. N. et al. Metal-independent variants of phosphoglycerate mutase promote resistance to nutritional immunity and retention of glycolysis during infection. PLoS Pathog.15, e1007971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nagpal, S. & Nair, D. T. The PHP domain of PolX from Staphylococcus aureus aids high fidelity DNA synthesis through the removal of misincorporated deoxyribo-, ribo- and oxidized nucleotides. Sci. Rep.11, 4178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reichert, S. et al. Genetic adaptation of a mevalonate pathway deficient mutant in Staphylococcus aureus. Front. Microbiol.9, 1539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kinkel, T. L., Roux, C. M., Dunman, P. M. & Fang, F. C. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio4, e00696–00613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schwan, W. R. & Wetzel, K. J. Osmolyte transport in Staphylococcus aureus and the role in pathogenesis. World J. Clin. Infect. Dis.6, 22–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou, C. et al. Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection. PLoS Pathog.15, e1007538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duval, B. D., Mathew, A., Satola, S. W. & Shafer, W. M. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob. Agents Chemother.54, 2283–2290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reslane, I. et al. Catabolic ornithine carbamoyltransferase activity facilitates growth of Staphylococcus aureus in defined medium lacking glucose and arginine. mBio13, e0039522 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhu, Y. et al. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun.75, 4219–4226 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Conlon, B. P. et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol.1, 16051 (2016). [DOI] [PubMed]
- 142.Shi, R. et al. Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase. Proc. Natl. Acad. Sci. USA108, 1302–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bennison, D. J. et al. The stringent response inhibits 70S ribosome formation in Staphylococcus aureus by impeding GTPase-ribosome interactions. mBio12, e0267921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schoenfelder, S. M. et al. Methionine biosynthesis in Staphylococcus aureus is tightly controlled by a hierarchical network involving an initiator tRNA-specific T-box riboswitch. PLoS Pathog.9, e1003606 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Potter, A. D. et al. Host nutrient milieu drives an essential role for aspartate biosynthesis during invasive Staphylococcus aureus infection. Proc. Natl. Acad. Sci. USA117, 12394–12401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Secor, P. R. et al. Phevalin (aureusimine B) production by Staphylococcus aureus biofilm and impacts on human keratinocyte gene expression. PLoS ONE7, e40973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lobo, S. A. et al. Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway. Mol. Microbiol.97, 472–487 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Chaudhari, S. S. et al. Nitrite derived from endogenous bacterial nitric oxide synthase activity promotes aerobic respiration. mBio8, e00887-17 (2017). [DOI] [PMC free article] [PubMed]
- 149.Singh, R. M. et al. A critical role for staphylococcal nitric oxide synthase in controlling flavohemoglobin toxicity. Redox Biol.67, 102935 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nguyen, T. et al. Targeting mannitol metabolism as an alternative antimicrobial strategy based on the structure-function study of mannitol-1-phosphate dehydrogenase in Staphylococcus aureus. mBio10, e02660-18 (2019). [DOI] [PMC free article] [PubMed]
- 151.Handke, L. D., Hawkins, J. C., Miller, A. A., Jansen, K. U. & Anderson, A. S. Regulation of Staphylococcus aureus MntC expression and its role in response to oxidative stress. PLoS ONE8, e77874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hall, J. W., Yang, J., Guo, H. & Ji, Y. The Staphylococcus aureus AirSR two-component system mediates reactive oxygen species resistance via transcriptional regulation of staphyloxanthin production. Infect. Immun.85, e00838-16 (2017). [DOI] [PMC free article] [PubMed]
- 153.Horsburgh, M. J. et al. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol.44, 1269–1286 (2002). [DOI] [PubMed] [Google Scholar]
- 154.Kaito, C., Kurokawa, K., Hossain, M. S., Akimitsu, N. & Sekimizu, K. Isolation and characterization of temperature-sensitive mutants of the Staphylococcus aureus dnaC gene. FEMS Microbiol. Lett.210, 157–164 (2002). [DOI] [PubMed] [Google Scholar]
- 155.Paudel, S., Bagale, K., Patel, S., Kooyers, N. J. & Kulkarni, R. Human urine alters methicillin-resistant Staphylococcus aureus virulence and transcriptome. Appl. Environ. Microbiol.87, e0074421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ballal, A. & Manna, A. C. Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus. J. Bacteriol.192, 336–345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ghssein, G. et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science352, 1105–1109 (2016). [DOI] [PubMed] [Google Scholar]
- 158.Brown, S., Zhang, Y. H. & Walker, S. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol.15, 12–21 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim, J., Kim, G. L., Norambuena, J., Boyd, J. M. & Parker, D. Impact of the pentose phosphate pathway on metabolism and pathogenesis of Staphylococcus aureus. PLoS Pathog.19, e1011531 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schiebel, J. et al. Staphylococcus aureus FabI: inhibition, substrate recognition, and potential implications for in vivo essentiality. Structure20, 802–813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Balibar, C. J., Shen, X. & Tao, J. The mevalonate pathway of Staphylococcus aureus. J. Bacteriol.191, 851–861 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Koprivnjak, T. et al. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J. Bacteriol.193, 4134–4142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jiang, J. H. et al. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. USA116, 3722–3727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen, C., Yang, C. & Barbieri, J. T. Staphylococcal superantigen-like protein 11 mediates neutrophil adhesion and motility arrest, a unique bacterial toxin action. Sci. Rep.9, 4211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Paharik, A. E. et al. The Spl serine proteases modulate Staphylococcus aureus protein production and virulence in a rabbit model of pneumonia. mSphere1, 10–1128 (2016). [DOI] [PMC free article] [PubMed]
- 166.Fisher, R. A., Gollan, B. & Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol.15, 453–464 (2017). [DOI] [PubMed] [Google Scholar]
- 167.Wong Fok Lung, T. et al. Staphylococcus aureus small colony variants impair host immunity by activating host cell glycolysis and inducing necroptosis. Nat. Microbiol.5, 141–153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Murdoch, C. C. & Skaar, E. P. Nutritional immunity: the battle for nutrient metals at the host-pathogen interface. Nat. Rev. Microbiol.20, 657–670 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Grayczyk, J. P., Harvey, C. J., Laczkovich, I. & Alonzo, F. 3rd A lipoylated metabolic protein released by Staphylococcus aureus suppresses macrophage activation. Cell Host Microbe22, 678–687 e679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zheng, X. et al. The cell envelope of Staphylococcus aureus selectively controls the sorting of virulence factors. Nat. Commun.12, 6193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Thurlow, L. R. et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol.186, 6585–6596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Van Roy, Z. et al. Tissue niche influences immune and metabolic profiles to Staphylococcus aureus biofilm infection. Nat. Commun.15, 8965 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fey, P. D. et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio4, e00537–00512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pang, Y. Y. et al. Agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immun.2, 546–559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yajjala, V. K. et al. Resistance to acute macrophage killing promotes airway fitness of prevalent community-acquired Staphylococcus aureus strains. J. Immunol.196, 4196–4203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Heim, C. E. et al. IL-12 promotes myeloid-derived suppressor cell recruitment and bacterial persistence during Staphylococcus aureus orthopedic implant infection. J. Immunol.194, 3861–3872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal17, 10–12 (2011).
- 178.Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res.27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Diep, B. A. et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet367, 731–739 (2006). [DOI] [PubMed] [Google Scholar]
- 180.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 182.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Perez, F. & Granger, B. E. IPython: a system for interactive scientific computing. Comput. Sci. Eng.9, 21–29 (2007). [Google Scholar]
- 184.Fuchs, S. et al. AureoWiki—the repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol.308, 558–568 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Sequencing data and processed count matrices have been deposited in the GEO database under accession code GSE270986. Source data are provided with this paper.