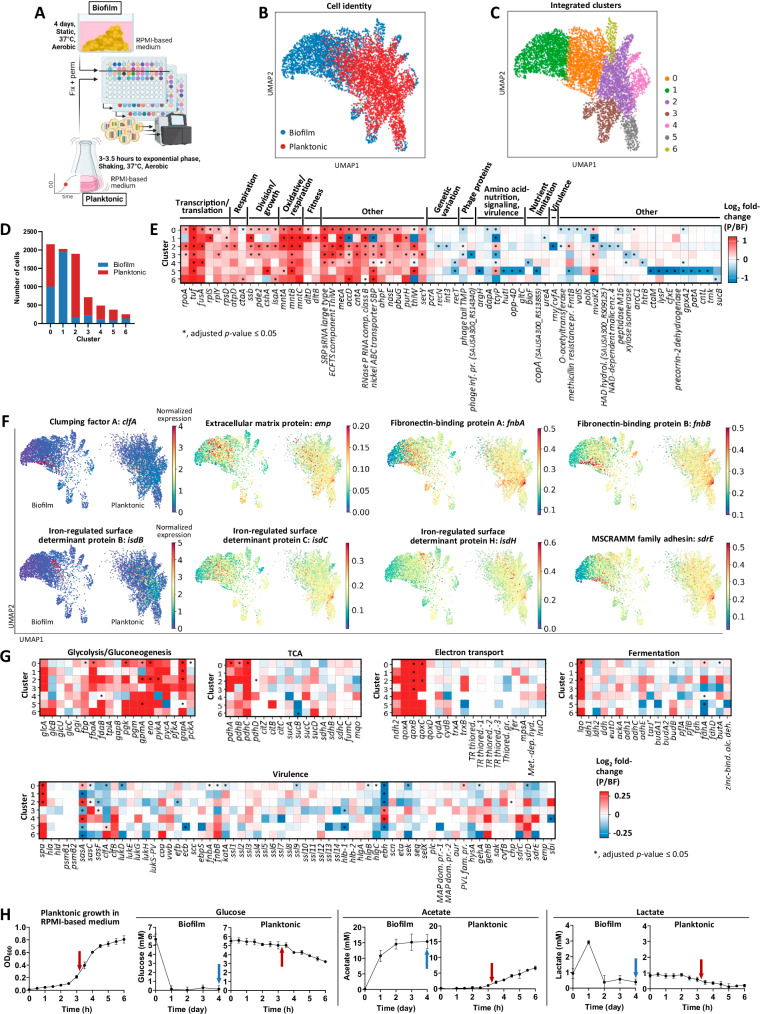
Fig. 2. Biofilm growth is marked by extensive transcriptional heterogeneity and decreased metabolic gene expression at the single-cell level.
A S. aureus biofilm was grown for 4 days under static conditions with daily medium replenishment, and planktonic culture was grown to exponential phase between 3 and 3.5 h with shaking at 250 rpm. Both biofilm and planktonic samples were grown in identical RPMI-based medium with aerobic incubation at 37 °C. Cells from biofilm and planktonic samples were fixed overnight before permeabilization under identical conditions. Cells from biofilm and planktonic cultures were separate for the first round of barcoding, then combined for the second and third rounds. The combined samples were processed through to sequencing. Schematic created in BioRender: Korshoj, L. (2024) BioRender.com/o01l597. B–C UMAP plots of the integrated biofilm and planktonic samples depicting (B) sample origin and (C) subpopulations identified with the Leiden algorithm. D Distribution of biofilm and planktonic cells across each cluster, reflecting 3680 cells for biofilm and 4231 cells for planktonic. E Marker genes specific to planktonic and biofilm growth across all clusters, represented as log2 fold-change of planktonic (P)/biofilm (BF). Red signifies upregulation in planktonic and blue signifies upregulation in biofilm. Additional genes are listed in Supplementary Data 1–7. False discovery rate (FDR)-adjusted p-values are noted. SRP signal recognition particle, ECFTS energy coupling factor transporter S, SBP substrate-binding protein, TPM tape measure protein. F Expression of exoproteome-associated genes overlaid on UMAP plots, separated by biofilm and planktonic conditions. Color represents normalized expression level on a per cell basis. G Comparisons of metabolic and virulence factor gene expression between planktonic and biofilm growth, represented as log2 fold-change of planktonic (P)/biofilm (BF). Red signifies upregulation in planktonic and blue signifies upregulation in biofilm. FDR-adjusted p-values are noted. TR thiol reductase, MAP MHC class II analog protein, TR thiored, SAUSA300_RS04260; TR thiored.−1, SAUSA300_RS04295; TR thiored.−2, SAUSA300_RS09235; TR thiored.−3, SAUSA300_RS13730; MAP dom. pr.−1, SAUSA300_RS10495; MAP dom. pr.−2, SAUSA300_RS10500. H Quantification of glucose, acetate, and lactate in culture supernatants collected during biofilm and planktonic growth (data are presented as mean ± standard deviation from 4 biological replicates). The planktonic growth curve in RPMI-based medium is also shown on the left (data are presented as mean ± standard deviation from 3 biological replicates). Arrows indicate the time of sample collection at which BaSSSh-seq was performed. Source data are provided as a Source Data file.