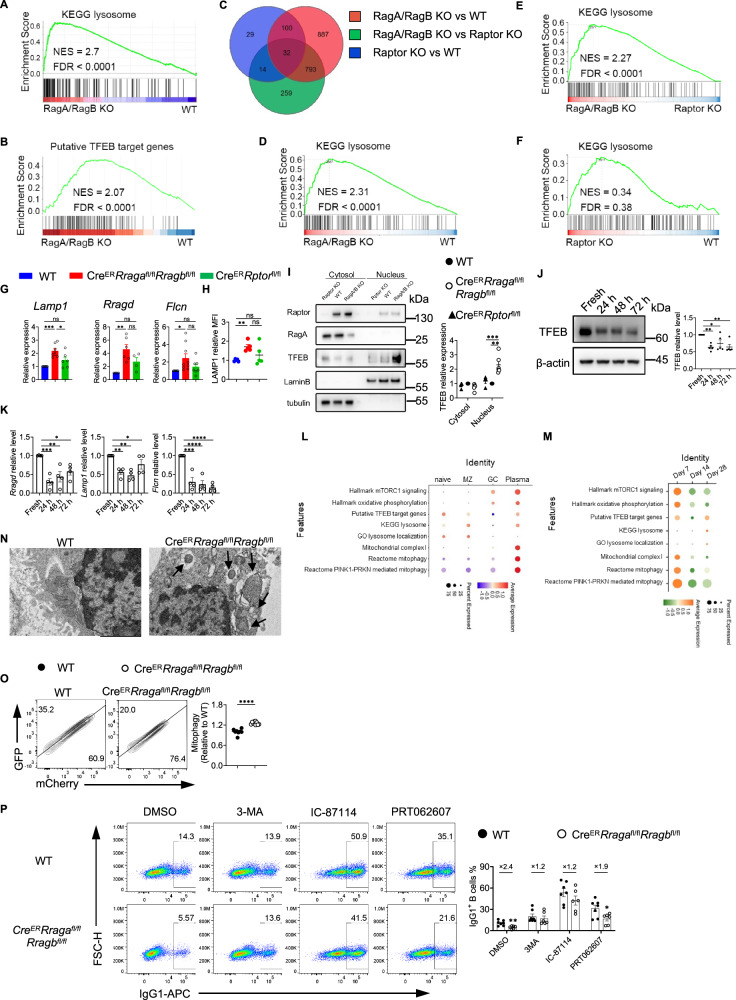
Fig. 5. RagA/RagB deficiency leads to TFEB/TFE3 overactivation and abnormal mitophagy.
A RNA sequencing was performed on 72-h activated B cells. Gene set enrichment analysis (GSEA) was conducted using differentially expressed genes (DEGs) with KEGG lysosome pathway plotted. B The enrichment of putative TFEB target genes. C RNA sequencing was performed on sorted germinal center (GC) B cells. Venn diagram of three DEG comparisons was presented. D–F GSEA was performed using the DEGs between indicated genotypes with KEGG lysosome pathway enrichment between indicated genotypes presented. G qRT-PCR of Lamp1, Rragd, and Flcn expression. H Flow cytometry of LAMP1 expression, n = 5 mice per group. I Immunoblot of Raptor, RagA, and TFEB expression. Cytosolic and nuclear proteins were isolated from activated B cells. Lamin-B, nuclear control, tubulin, cytosol control. WT (n = 5), CreERRragafl/flRragbfl/fl (n = 7), and CreERRptorfl/fl (n = 4). J Expression of TFEB at indicated time-points (n = 5 for each condition). Right, summary of relative TFEB expression (normalized to fresh). K qRT-PCR of Lamp1, Rragd, and Flcn expression at indicated time-points. n = 4 for each condition. L, M Published scRNAseq dataset (E-MTAB-9478) was reanalyzed with gene signatures in naïve, marginal zone (MZ), GC B cells, and plasma cells presented (L). Gene signatures in GC B cells at different time points (M). N Transmission electron microscopy (TEM) of activated B cells. Arrows indicate mitochondria surrounded by double-layer structures. Scale bar, 2 μm. O GFP and mCherry expression with Mito-QC transduction. Right, summary of the normalized mCherry percentages (Relative to mCherry percentage in WT). WT (n = 8), and CreERRragafl/flRragbfl/fl (n = 10). P IgG1 expression on B cells stimulated in the presence of indicated inhibitors. Right, summary of IgG1+ B cell percentages. WT (n = 7), CreERRragafl/flRragbfl/fl (n = 6). Data represents at least 3 (G–K, O, P) and 2 (N) independent experiments. Data in graphs represent mean ± SEM. ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, one-way ANOVA (G–K), two-tailed/unpaired Student’s t-test (O and P). Source data are provided as a Source Data file.