Abstract
Early-onset colorectal cancer (EOCRC), recognized as a distinct subgroup with an increased incidence over the past two decades, characterized by its aggressive nature and potentially unique molecular factors that differentiate it from traditional colorectal cancer (CRC). In this study, we investigated differentially expressed genes in a young-CRC patient using paired-end mRNA-sequencing. Validation of target genes through qRT-PCR highlighted a significant increase in SERPINA3 levels in EOCRC, representing a novel finding. Epithelial expression of SERPINA3 demonstrated a strong association with disease progression, whereas stromal expression showed a negative correlation. Our findings reveal the distinct expression patterns and potential involvement of SERPINA3 in both the initiation and progression of CRC, suggesting that SERPINA3 could serve as a marker for distinguishing early-onset from late-onset cases.
Keywords: Early-onset of colorectal cancer, SERPINA3, Inflammation, Stroma, Epithelial cell, Colorectal cancer
1. Introduction
Colorectal cancer (CRC) ranks second in cancer-related mortality and third in incidence Worldwide [1,2]. Over the last two decades, the escalating incidence of CRC in individuals under 50 years of age has become a global and particularly concerning trend in India [2]. Although screening has significantly contributed to early CRC diagnosis and prevention, it conventionally targets individuals between 50 and 60 years old, raising concerns about its efficacy in the younger CRC population. Early-onset CRC (EOCRC) tends to manifest at advanced disease stages [3], and comprises a notable proportion of sporadic cases [4]. While the aetiology of EOCRC lacks a definitive explanation, risk factors such as inflammatory bowel disease (IBD), gut dysbiosis, race, gender, and family history of CRC have been implicated [5,6]. A large case-control study including EOCRC, late-onset CRC (LOCRC), and healthy controls revealed a significantly higher prevalence of IBD in EOCRC patients compared to healthy controls, with IBD patients having nearly a threefold higher risk and frequency of developing EOCRC than late-onset CRC [6,7]. Few studies have suggested that sporadic EOCRC differs in its pathological and molecular nature, with an elevated immune signature compared to late-onset cases [8,9]. Inflammation and immune response activation have been implicated in the onset of EOCRC. Notably, molecular distinctions between early and late onset CRC have been reported, with EOCRC often following non-canonical cascades [[10], [11], [12]], mostly originating in the sigmoid colon and rectum without predisposition [13]. Despite being considered a distinct subset of CRC due to these differences [9,14], the molecular disparities between young and traditional CRC remain underexplored.
During inflammation, the liver releases acute phase proteins (APPs), including serpins like α1-antitrypsin (AAT) and α1-antichymotrypsin (AACT), into the bloodstream, playing vital roles in immune response regulation and negative feedback on inflammation [15]. Serpins, the serine protease inhibitor superfamily members, are renowned for inhibiting serine proteases, with emerging evidence implicating their role in various diseases [16]. Particularly, AACT or SERine Protease INhibitor A3 (SERPINA3) has gained attention for altered expression linked to poor prognosis in cancers and potential associations with other diseases [17,18]. Predominantly activated by inflammatory pathways, SERPINA3 exhibits elevated levels in sporadic desmoid tumors and CRC, influencing invasion, migration, and liver metastases [[19], [20], [21]]. Noteworthy, in glioma tissues, heightened SERPINA3 correlates with suppressed CD4+ T cell infiltration, suggesting a role in immune suppression within the glioma microenvironment [22,23]. Despite its potential in tumor microenvironment (TME)-mediated cancer development, extensive research on SERPINA3's molecular mechanisms in physiology or pathology remains unexplored.
Indeed, understanding the molecular signatures associated with EOCRC could provide insights into the disease onset and open new avenues for early diagnosis and treatment. In this study, we explored differentially expressed genes in an EOCRC patient using paired-end mRNA sequencing and identified elevated SERPINA3 expression in EOCRC compared to LOCRC. SERPINA3 exhibited differential expression in the epithelial and stromal components of rectal tissue arrays. Additionally, the differential localization of SERPINA3 in tumor epithelia across different cohorts was observed, adding an intriguing dimension to its role in EOCRC.
2. Materials and methods
2.1. Collection of biopsy samples
Biopsy specimens were acquired from patients who underwent adjuvant or neo-adjuvant therapy for CRC at Govt. Medical College, Kollam, Kerala, India following informed consent and ethical approval (IHEC/1/2018/05). Tumor and adjacent normal tissue samples from the resected colon or rectum were individually preserved in RNAlater tubes (Invitrogen, USA). The study exclusively utilized paired tumor samples along with corresponding adjacent matched normal samples. The tissues were promptly processed for total RNA isolation or stored at −80 °C for subsequent RNA extraction. Additionally, some specimens were used to create paraffin blocks for immunohistochemistry analysis.
2.2. RNA isolation
Total RNA was extracted from biopsies using TRIzol reagent (Ambion, USA). Approximately 50 mg of tissue was mechanically homogenized in 1 mL of TRIzol, followed by phase separation using chloroform. The aqueous phase was transferred to a new tube, and 500 μL of isopropanol was added for RNA precipitation. The RNA pellet underwent two washes with 75 % ethanol, followed by air-drying for 30 min. Subsequently, the RNA pellet was reconstituted in nuclease-free water for further analysis. The concentration and purity of the isolated total RNA were assessed using a Nanodrop ND-1000 spectrophotometer.
2.3. Quantitative real-time-PCR
Quantitative real-time PCR (qRT-PCR) was conducted using the SYBR-Green-based fluorescence detection method and the HT9700 detection system (AB, Life Science, USA). Primers for the study were designed, and their specificity was determined through BLAST analysis using the NCBI primer designing tool (Table S1, primers were purchased from Integrated DNA Technologies, USA). Total RNA underwent reverse transcription using the PrimeScript First Strand cDNA Synthesis Kit (TaKaRa, Japan). SERPINA3 mRNA levels were quantified in both tumor and respective normal samples by qRT-PCR using SYBR Premix Ex Taq II (TaKaRa, USA) and conducted on the 7900HT Fast Real-Time PCR System (Applied Biosystems, USA). The relative expressions of genes were determined by analysing the Log2 fold change of gene expression in tumor samples using the ΔΔCT method (Data Assist software v3.01, AB Life Science, USA). Log2 fold change was obtained by normalizing normal and tumor samples from each patient to the housekeeping gene L19 as the loading control before normalizing the fold change of the tumor to their respective normal sample. Fold change as well as Log2 fold change values were used for result analysis.
2.4. High throughput RNA sequencing and differential gene expression analysis
Oligo(dT) beads were employed for mRNA purification, followed by RNA fragmentation. The first strand of cDNA was synthesized using random hexamer primers, and the second strand was generated using DNA polymerase, dNTPs, RNase H, and buffer. DNA purification occurred using magnetic beads, followed by ligation of sequencing adaptors to fragments and PCR amplification. Sequencing was conducted using Illumina Next Seq 500. Raw reads underwent quality checks with FastQC. An in-house script removed adaptors and trimmed low-quality reads towards the 3′ end. TopHat-2.0.7 aligned RNA-Seq reads to mammalian-sized genomes via the ultra-high-throughput short read aligner, Bowtie, identifying splice junctions. Cufflinks 2.0.1 assembled transcripts, estimated their abundance, and assessed differential expression and regulation. Cuffdiff tool was utilized for analysing differential gene expression.
2.5. Gene function and pathways enrichment analysis
The Log2 fold change and p-value criteria were employed to filter significantly altered genes in the tumor compared to normal tissue. Subsequently, the DAVID bioinformatics tool (https://david.ncifcrf.gov/) was used for further analysis of these significantly altered genes. The tool facilitated gene ontology (GO) analysis, categorizing genes into bioprocess, cellular components, and molecular functions. It also allowed the examination of altered signalling cascades from the input gene list. To identify genes associated with a specific GO process, the AmiGO 2 software (http://amigo.geneontology.org/amigo) was utilized. Our specific query focused on the GO term associated with the Maintenance of Gastrointestinal Epithelium -MGE (GO:0030277) and Inflammatory Response Bioprocess -IRB (GO:0006954) in the AmiGO 2 database. The resulting output provided genes associated with these GO terms, which were then cross-referenced with our Differential Gene Expression (DGE) list using the Bioinformatics & Evolutionary Genomics webtool (https://bioinformatics.psb.ugent.be/webtools/Venn/) to identify common genes.
2.6. Histopathological staining
Following surgery, tissues were fixed using 4 % paraformaldehyde (Sigma-Aldrich, USA) for 16 h at 4 °C. Subsequently, the tissues underwent processing with xylene and alcohol gradient, followed by embedding in paraffin to create paraffin blocks. Sections of 5 μm thickness were mounted on poly L-lysine-coated StarFrost glass slides (Leica, Germany). Deparaffinization utilized xylene (SRL, India), rehydration involved isopropyl alcohol (Merck, USA), and H2O2. Rehydrated sections were treated with hematoxylin and incubated at room temperature for 3 min, followed by an acid alcohol dip. Post-incubation, slides were washed with water and treated with bluing Scott's solution for 10 min. After bluing, slides were rinsed with water, dehydrated with 70 % isopropyl alcohol, and counter-stained with eosin. The counter-stained sections were rinsed with alcohol gradient and xylene for dehydration. Finally, the slides were mounted and observed under an upright microscope (Leica DM1000, Germany).
2.7. Immunohistochemistry (IHC) analysis
Tissues obtained post-surgery underwent fixation with 4 % paraformaldehyde (Sigma-Aldrich, USA) at 4 °C for 16 h. Subsequently, tissues were processed using an alcohol gradient and xylene, followed by embedding in paraffin to create paraffin blocks. Sections of 5 μm thickness were mounted on StarFrost glass slides (Leica, Germany). Deparaffinization involved three changes of xylene, followed by rehydration using a series of alcohol gradients to water. Sections were subjected to endogenous peroxidase blocking, and antigen retrieval was performed using Tris-EDTA buffer (pH 9). After retrieval, peroxidase quencher (H2O2) blocking was carried out for 5 min at room temperature, followed by three 1X PBST washes. Tissue boundaries were delineated using DPX Mountant (SRL, India) and allowed to dry. Sections were then incubated overnight at 4 °C in a humid chamber with the SERPINA3 primary antibody (dilution 1:1000, Abcam, USA). After each incubation, the tissue was washed with 1X PBST. The tissue underwent incubation with poly-target binder and poly-HRP for 15 min, followed by washes. DAB substrate (Sigma-Aldrich, USA) served as the chromogen, and hematoxylin (Merck Millipore, Germany) was used as a counterstain. Sections without the primary antibody incubation served as negative controls. The aforementioned IHC methodology was also applied to rectal tissue microarray (TMA) slides RE804 and REC2281, which were purchased from US Biomax, USA.
2.8. Immuno-reactive score (IRS) calculation
Both the TMA and slides were made using the samples collected from our cohort were examined using an upright microscope (Leica DM1000, Germany). For scoring, three distinct random fields were selected from each section. Interpretation encompassed independent assessments of cytoplasmic and nuclear staining for epithelial and stromal cells and the representative figures shown in Fig. S1. A pathologist supervised the determination of intensity (I) and the count of positive cells/area positivity (P) for each field, and the IR-score was manually calculated, as detailed in Table S2 [24]. The product of P and I from each field was averaged (P∗I), and the resulting values from the three fields determined the final IR-scores for each sample. IR-scores ranged from 0 to 12, categorized into SERPINA3 low (IR-scores <3) and high (IR-scores ≥3) expression groups. These IR-scores were utilized for graph plotting, and the categorized groups were employed for comparing SERPINA3's association with variables using SPSS statistical analysis software.
2.9. Statistical analysis
All collected data underwent analysis using statistical tools, including IBM SPSS Statistics 20 (SPSS Inc., Chicago, USA) and GraphPad Prism 9 (GraphPad Software Inc., California, USA). The unpaired Student's t-test was applied to compare means of two independent groups, while the paired t-test was utilized for dependent groups, with results expressed as mean ± standard error mean (SEM). For binomial variables, the Chi-square test and Fisher's exact test were employed. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001 were considered statistically significant.
3. Results
3.1. SERPINA3 is highly expressed in EOCRC and associated with poor survival
To uncover the gene expression profile characteristic of EOCRC, we conducted RNA sequencing on a CRC patient in the T4 stage at the age of 36, utilizing the Illumina Next Seq 500 platform (Genotypic Technology, Bangalore, India) [25]. Fig. 1A shows graphical representation of the workflow illustrating the high-throughput analysis, identification, and validation of target gene(s). A matched normal sample served as a reference for gene expression comparison. The analysis yielded a total of 27,905,573 and 24,083,677 processed reads for the normal and tumor samples, respectively (Table S3).
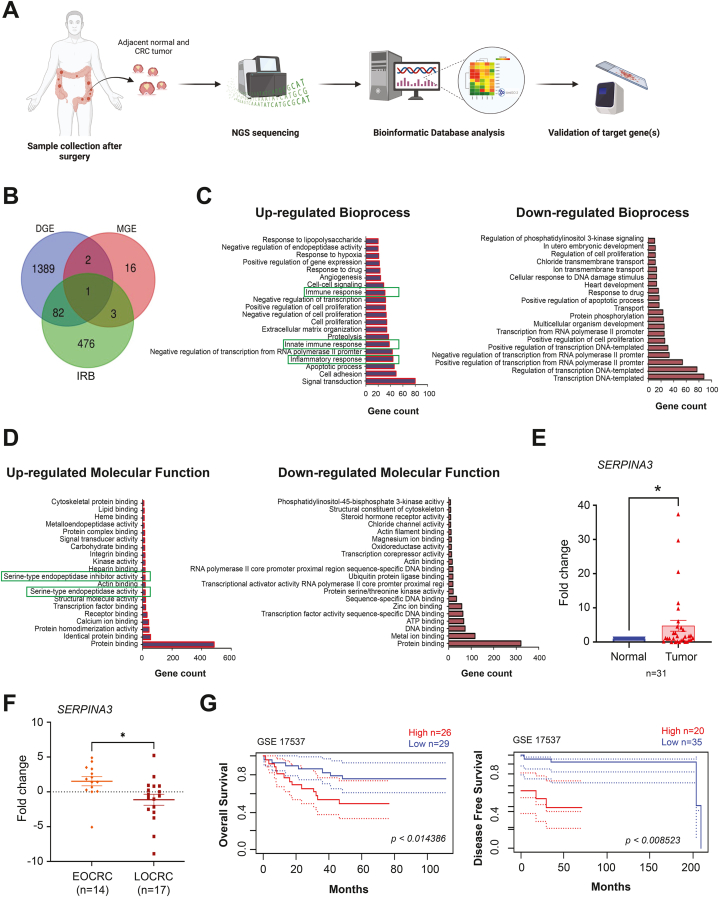
Fig. 1.
SERPINA3 is highly expressed in early onset CRC and associated with poor survival. A Schematic workflow of the study shows the high-throughput analysis, identification and validation of target gene/s. B Venn diagram showing genes enriching maintenance of gastrointestinal epithelium (MGE) bioprocess, inflammatory response bioprocess (IRB) and differential gene expression (DGE) datasets. C-D Top 20 upregulated and downregulated bioprocess and molecular functions retrieved from DAVID database analysis of the deregulated genes. E Real-time q-PCR data showing expression of SERPINA3 in CRC compared to the matched normal biopsies. F SERPINA3 mRNA expression in early onset and late onset of CRC patient tissues measured by real-time PCR. G Kaplan–Meier plots showed high expression of SERPINA3 is associated with a decrease in overall and disease-free survival in CRC patients. Data is presented as the mean ± SEM. ∗p < 0.05 consider statistical significance.
Differential gene expression (DGE) analysis identified 816 upregulated and 658 downregulated genes, meeting the criteria of log2 fold change ±1.5 and a p-value ≤0.05 (Fig. 1, B). Genes implicated in intestinal development play a pivotal role in upholding enteric cells, and any genomic or expression alterations in these genes could potentially contribute to tumorigenesis. Currently, there is a notable dearth of discussion defining altered gene signatures in EOCRC. Consequently, we aimed to pinpoint the genes associated with gastrointestinal development that exhibit differential expression in early CRC. To achieve this, we overlaid gene ontology for maintenance of gastrointestinal development (MGE) onto our set of DGE (Fig. 1, B). Notably, three genes—VSIG1 (log2 fold change 5.45), SERPINA3 (log2 fold change 2.87), and TFF1 (log2 fold change 1.96)—emerged as significantly upregulated with a p-value <0.05. Subsequently, we delved into the genes enriching inflammatory response bioprocess (IRB) and identified SERPINA3 as the distinct gene in our list associated with MGE and IRB (Fig. 1, B). Subsequently, the lists of altered genes were subjected to analysis for their biological and functional significance using the DAVID bioinformatics tool (Fig. 1C–D). Intriguingly, among the upregulated bioprocesses, we observed innate immune response and inflammatory immune response (Fig. 1, C), while serine-type endopeptidase inhibitor activity was noted among the upregulated molecular functions (Fig. 1, D). Therefore, SERPINA3 was chosen for further in-depth study.
To validate SERPINA3 expression in clinical samples, paired tumor and adjacent normal tissues were obtained from patients for the study. Patient characteristics are summarized in Table 1. qPCR was employed to assess gene expression in these samples, revealing a significantly elevated SERPINA3 mRNA expression in CRC compared to matched normal tissues (Fig. 1, E). Subsequently, we stratified our cohort based on age, distinguishing between early-onset (≤50 years) and late-onset (>50 years) groups. A notable difference in SERPINA3 expression levels emerged between these two groups. Specifically, SERPINA3 is overexpressed (log2 fold change 1.528) exclusively in the EOCRC in contrast to the LOCRC, where it exhibited downregulation (log2 fold change −1.131) (Fig. 1, F). In order to evaluate the impact of SERPINA3 expression on the survival of CRC patients, we analysed publicly available dataset (GEO accession: GSE17537) for SERPINA3 expression using the PrognoScan platform (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html). Kaplan-Meier analysis revealed a significant decrease (p < 0.01 and p < 0.008) in both overall and disease-free survival among patients with elevated SERPINA3 expression, indicating its potential as a promoter of tumor progression in CRC patients (Fig. 1, G).
Table 1.
Patient characteristics were used in the present study.
| Total number of patients 53 | ||
|---|---|---|
| Age (years) | <50 | 20 |
| ≥50 | 32 | |
| Not known | 1 | |
| Sex | Women | 26 |
| Men | 26 | |
| Not known | 1 | |
| Treatment | Pre-CTRT | 24 |
| Post-CTRT | 26 | |
| Unknown | 3 | |
| Depth of Invasion | ≤p T2 | 8 |
| >p T2 | 18 | |
| Not known | 27 | |
| Lymph node metastasis | Negative | 9 |
| Positive | 15 | |
| Not known | 29 | |
| Distant metastasis | Negative | 20 |
| Positive | 4 | |
| Not known | 29 | |
| Histologic type | Well differentiated | 9 |
| Moderately differentiated | 19 | |
| Poorly differentiated | 1 | |
| Not known | 24 | |
| Site of tumor | Colon | 8 |
| Rectum and sigmoid | 39 | |
| Not known | 6 | |
| Samples used for the study | RT-PCR | 31 |
| IHC | 23 | |
| Common sample/s | 1 | |
CRT=Chemoradiotherapy, RT-PCR=Real time PCR, IHC=Immunohistochemistry.
3.2. Association of SERPINA3 levels in epithelial and stromal cells with the progression of disease
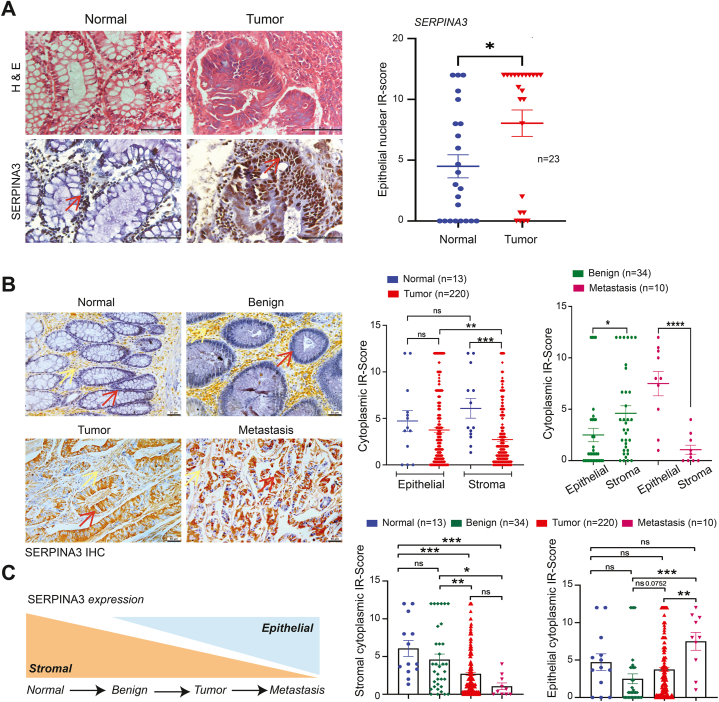
In this investigation, we delve into the pathological implications of SERPINA3 to comprehend its role in EOCRC and the initial stages of cancer development. IHC analysis of patient samples was conducted to elucidate the association of SERPINA3 throughout the progression of CRC, spanning from the normal stage to metastasis. SERPINA3 exhibited expression in the cytoplasm and nucleus of both epithelial and stromal cells in CRC specimens, prompting the consideration of separate and combined scores for nuclear and cytoplasmic staining in epithelial and stromal cells. Epithelial cells predominantly displayed nuclear staining with varying degrees of cytoplasmic staining in tumor and normal tissues. Conversely, stromal cells in normal and tumor tissues exhibited comparable nuclear and cytoplasmic staining. Tumor epithelium demonstrated significantly higher nuclear SERPINA3 levels compared to matched normal tissues (Fig. 2, A). However, stromal cytoplasmic or nuclear SERPINA3 expression in tumors did not exhibit statistical differences compared to matched normal samples. Limited tumor sections displayed SERPINA3 cytoplasmic epithelial staining, precluding quantification. However, epithelial and stromal SERPINA3 expression did not show any statistical association with age or sex (Table 2).
Fig. 2.
Association of SERPINA3 levels in epithelial and stromal tissues with the progression of disease.A Representative H&E and immunohistochemical (IHC) staining for SERPINA3 in normal and CRC colorectal tissues. Brown staining indicates SERPINA3. Scale bar = 20 μm. B Representative IHC images for SERPINA3 expression in progressive stages of rectal cancer of tissue microarray (normal, benign, tumor and metastasis). Scale bar = 50 μm. Red arrows indicate epithelial cells, while the yellow arrow highlights the stroma. C Consolidated summary graphs showing the epithelial and stromal cytoplasmic SERPINA3 expression to progressive stages of rectal cancer. Data is presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 consider statistical significance. ns = non-significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Association of SERPINA3 expression with the clinical and pathological parameters in CRC samples using SPSS statistical analysis.
| Variable | Total no. of cases | Epithelial staining (Nuclear) |
Stromal staining (Nuclear) |
Stromal staining (Cytoplasmic) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | high | p-value | Low | High | p-value | Low | High | p-value | |||
| Normal | 23 | 14 | 9 | 0.075a | 18 | 5 | 0.514a | 21 | 2 | 1.000a | |
| Tumor | 23 | 7 | 16 | 15 | 8 | 22 | 1 | ||||
| Adenocarcinoma | 23 | ||||||||||
| Age (years) | <50 | 6 | 2 | 4 | 1.000a | 5 | 1 | 1.000a | 6 | 0 | 1.000a |
| ≥50 | 16 | 4 | 12 | 13 | 3 | 15 | 1 | ||||
| aNA | 1 | ||||||||||
| Sex | Female | 9 | 2 | 7 | 1.000a | 8 | 1 | 0.616a | 8 | 1 | 0.409a |
| Male | 13 | 4 | 9 | 10 | 3 | 13 | 0 | ||||
| aNA | 1 | ||||||||||
| Organ | Colon | 4 | 0 | 4 | 0.245a | 3 | 1 | 1.000a | 4 | 0 | 1.000a |
| Rectum | 14 | 6 | 8 | 11 | 3 | 13 | 1 | ||||
| aNA | 5 | ||||||||||
| Treatment | Primary | 15 | 1 | 14 | 0.001a | 11 | 4 | 0.263a | 15 | 0 | 0.318a |
| Post-CTRT | 7 | 6 | 1 | 7 | 0 | 6 | 1 | ||||
| aNA | 1 | ||||||||||
| Stage | I&II | 5 | 1 | 4 | 0.143a | 4 | 1 | 1.000a | 4 | 1 | 1.000a |
| III&IV | 3 | 3 | 0 | 3 | 0 | 3 | 0 | ||||
| aNA | 15 | ||||||||||
| Grade | WD | 3 | 1 | 2 | 1.000a | 2 | 1 | 0.333a | 3 | 0 | 1.000a |
| MD | 6 | 3 | 3 | 6 | 0 | 5 | 1 | ||||
| aNA | 14 | ||||||||||
NA = not available, a = F-test.
As per certain studies [2,26], EOCRC is more commonly associated with rectal cancer than colon cancer. While CRC was traditionally regarded as a single entity, recent investigations have unveiled distinct disparities between rectal and colon cancer concerning molecular carcinogenesis, pathology, surgical topography, techniques, and multimodal therapy [27]. Considering the notion that colon and rectal cancer are deemed distinct malignancies, and EOCRC is reported to exhibit rectal predominance, we examined the expression of SERPINA3 at various stages of rectal cancer to explore its involvement in cancer initiation and progression. Tissue arrays with a total of 300 samples were analysed for the study, and detailed patient characteristics are provided in Table 3. IHC analysis of tissue arrays (US cohort) revealed predominant cytoplasmic staining of SERPINA3 in both epithelial and stromal cells (Fig. 2, B), in contrast to the predominant nuclear SERPINA3 staining in epithelia from the Indian cohort (Fig. 2, A). Epithelial and stromal SERPINA3 expression did not show any statistical association with age or sex (Table 4).
Table 3.
Patient characteristics table from tissue arrays purchased from US Biomax.
| Patients characteristics | ||
|---|---|---|
| Total number of samples | 300 | |
| Age (years) | >50 ≤50 |
97 203 |
| Sex | Women Men |
107 193 |
| Depth of Invasion | ≤p T2 | 105 |
| >p T2 | 138 | |
| Lymph node metastasis | Negative | 154 |
| Positive | 89 | |
| Distant metastasis | Negative | 241 |
| Positive | 2 | |
| Location of tumor | Rectum | 290 |
| Lymph node | 9 | |
| Mesentery | 1 | |
| Histologic type | Well differentiated | 60 |
| Moderately differentiated | 147 | |
| Poorly differentiated | 28 | |
| Pathology at diagnosis | Normal adjacent to rectal cancer tissue (NAT) | 13 |
| Polyp | 4 | |
| Adenoma | 15 | |
| Hyperplasia | 10 | |
| Inflammation | 5 | |
| Adenocarcinoma | 220 | |
| Other | 23 | |
| Metastatic adenocarcinoma of mesentery from rectum | 10 | |
Table 4.
Association of SERPINA3 expression with the clinical and pathological parameters using SPSS statistical analysis from rectal cancer tissue array samples.
| Variable |
Total no. of cases |
Epithelial staining (Cytoplasmic) |
Stromal staining (Cytoplasmic) |
|||||
|---|---|---|---|---|---|---|---|---|
| Low | high | p-value | Low | High | p-value | |||
| Adenocarcinoma | 220 | |||||||
| Age (years) | <50 | 62 | 33 | 29 | 0.882a | 37 | 25 | 0.207a |
| ≥50 | 158 | 86 | 72 | 109 | 49 | |||
| Sex | Female | 147 | 80 | 67 | 1.000a | 103 | 44 | 0.129a |
| Male | 73 | 39 | 34 | 43 | 30 | |||
| Stage | I&II | 137 | 69 | 68 | 0.165a | 91 | 46 | 1.000a |
| III&IV | 83 | 50 | 33 | 55 | 28 | |||
| Grade | I | 57 | 35 | 22 | 0.417b | 41 | 16 | 0.229b |
| II | 141 | 72 | 69 | 88 | 53 | |||
| III | 22 | 12 | 10 | 17 | 5 | |||
| T | I | 5 | 5 | 0 | 0.020b | 4 | 1 | 0.889b |
| II | 87 | 48 | 39 | 56 | 31 | |||
| III | 112 | 62 | 50 | 75 | 37 | |||
| IV | 16 | 4 | 12 | 11 | 5 | |||
| Lymph node status | Negative | 138 | 69 | 69 | 0.125a | 91 | 47 | 0.884a |
| Positive | 82 | 50 | 32 | 55 | 27 | |||
= F-test.
= Chi-square test.
Initially, we examined the difference in expression levels of SERPINA3 in normal (n = 13) vs. tumor samples (n = 220) (Fig. 2, B). Cytoplasmic SERPINA3 expression in tumor epithelia did not show a significant difference compared to normal epithelia, while cytoplasmic SERPINA3 in normal stroma showed significant upregulation compared to tumor stroma. Epithelial cytoplasmic SERPINA3 levels were significantly elevated compared to stromal levels in rectal adenocarcinoma samples, while in normal samples, no significant difference in expression between epithelia and stroma was observed. Cytoplasmic SERPINA3 expression was significantly higher in stroma compared to epithelia in benign samples (n = 34) (Fig. 2, B). Conversely, cytoplasmic SERPINA3 was significantly elevated in epithelia compared to stroma in metastatic samples (n = 10). Cytoplasmic SERPINA3 expression for epithelia and stromal cells was scored for progressive stages, i.e., i) normal, ii) benign, iii) tumor, and iv) metastasis. Interestingly, epithelial cytoplasmic SERPINA3 showed an increasing trend from benign to malignant stages, even though normal cells exhibited elevated levels of SERPINA3. Stromal cytoplasmic SERPINA3 showed a decreasing trend in expression levels from normal to metastasis stages (Fig. 2, C).
3.3. SERPINA3 displays unique staining in the stroma of mucosal chronic inflammation contrary to rectal adenocarcinoma tissues
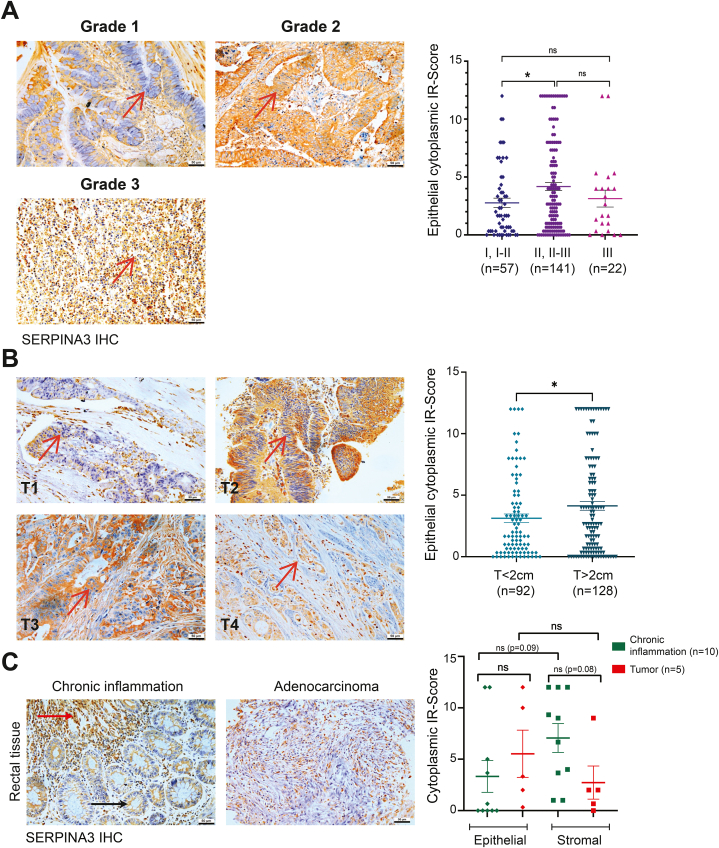
Rectal adenocarcinoma samples (n = 220) from the TMA were used to evaluate the association of SERPINA3 expression with cancer grades and invasive stages. We analysed the significance of epithelial and stromal cytoplasmic SERPINA3 levels in relation to the stage, grade, and TNM status of rectal adenocarcinomas. The Chi-square test revealed a significant association of epithelial cytoplasmic SERPINA3 expression with T stages of rectal adenocarcinomas (Table 4). SERPINA3 epithelial expression was significantly elevated in moderately differentiated rectal adenocarcinomas compared to well-differentiated samples (Fig. 3, A). Also, epithelial SERPINA3 showed significantly higher expression in T > 2 stages compared to T ≤ 2 stages of rectal adenocarcinoma (Fig. 3, B). However, no significant difference was found among T stage and grade status for stromal SERPINA3 expression in rectal adenocarcinomas.
Fig. 3.
SERPINA3 displays unique staining in the stroma of mucosal chronic inflammation contrary to rectal adenocarcinoma tissues. A-B Representative images and graph showing SERPINA3 expression in grades and T-stages of rectal tumors. Scale bar = 50 μm. Red arrows indicate epithelial cells. C Representative images and graph showing SERPINA3 expression in chronic inflammation of rectal mucosa (n = 10) and its respective rectal adenocarcinoma samples (n = 5, since paired samples for the remaining five were not available). Scale bar = 50 μm. Red arrow indicates nuclear and black arrow indicates cytoplasmic SERPINA3 staining. Data is presented as the mean ± SEM. ∗p < 0.05 consider statistical significance. ns = non-significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Given SERPINA3's pro-inflammatory characteristics and the established link between inflammation and CRC, we investigated the correlation of SERPINA3 in sections of chronic inflammation in rectal mucosa. We compared this expression with corresponding adenocarcinoma sections from the tissue array, assessing its presence in both the epithelial and stromal components of each tissue type. Upon analysis of cytoplasmic staining, elevated SERPINA3 expression was evident in the stroma of inflammatory samples compared to other groups (Fig. 3, C). Notably, only one out of five inflammatory samples exhibited high SERPINA3 epithelial expression, while all five paired malignant samples displayed positivity for epithelial staining. Analysis revealed a significant negative association for epithelial SERPINA3 staining between chronic inflammation and paired malignant samples (Table 5). Furthermore, three out of five inflammatory samples showed high SERPINA3 stromal expression, suggesting upregulation compared to epithelial SERPINA3 in chronic inflammatory samples. Interestingly, stromal SERPINA3 also appeared to be upregulated in chronic inflammatory samples compared to their paired adenocarcinomas. However, additional paired samples are required for a conclusive understanding of SERPINA3's localized expression and its association with inflammation and malignancy. Intriguingly, one chronic inflammation sample exhibited nuclear localization of SERPINA3 in epithelial cells progressing to higher stages or invading epithelia, while cytoplasmic staining in the epithelial cells remained in intact epithelia or lower stage tumors (Fig. 3, C).
Table 5.
SPSS analysis for the association of epithelial and stromal SERPINA3 expression on adenocarcinoma and chronic inflammation of rectal mucosa samples.
| Variable | Epithelial cytoplasmic SERPINA3 staining |
Stromal cytoplasmic SERPINA3 staining |
||||||
|---|---|---|---|---|---|---|---|---|
| N | Low | High | p-value | N | Low | High | p-value | |
| Rectal adenocarcinoma | 5 | 2 | 3 |
0.524a |
5 | 4 | 1 |
0.524a |
|
Chronic inflammation of rectal mucosa |
5 |
4 |
1 |
5 |
2 |
3 |
||
| Epithelial cytoplasmic SERPINA3 staining | Stromal cytoplasmic SERPINA3 staining | |||||||
| n |
No |
Yes |
p-value |
n |
No |
Yes |
p-value |
|
| Rectal adenocarcinoma | 5 | 0 | 5 | 0.048a | 5 | 1 | 4 | 1.00a |
| Chronic inflammation of rectal mucosa | 5 | 4 | 1 | 5 | 2 | 3 | ||
= F-test.
4. Discussion
EOCRC has become a global concern, with an increasing incidence reported in the past two decades [2]. Despite extensive research on CRC molecular signatures, limited attention has been given to EOCRC, which lacks a clear genetic predisposition [13]. Our study aimed to explore the altered gene expression in EOCRC using mRNA sequencing, focusing on genes related to gastrointestinal development and associated with MGE revealed SERPINA3, TFF1, and VSIG1 as significantly altered genes.
Chronic inflammation is a crucial CRC risk factor, and studies suggest its role in EOCRC [7]. Recent studies indicate that sporadic EOCRC differs in both pathology and molecular nature, with inflammatory signalling driving the disease [28]. The risk of developing EOCRC is notably higher in patients with IBD [7]. Inflammatory pathways have been reported to initiate carcinogenesis by activating different oncogenic cascades [29]. The AmiGO database and inflammatory response GO analysis identified SERPINA3 as the sole common gene among the altered genes associated with the MGE gene list. Due to its enrichment in the IRB from the altered gene list and considering the significance of inflammation in CRC development, we focused on SERPINA3. This serine protease inhibitor, known for its diverse roles in diseases such as Alzheimer's, prion diseases, and various cancers [17] was found to be up-regulated specifically in EOCRC, suggesting a potential molecular distinction between early and late onset CRC entities.
The onset of EOCRC is hypothesized to be linked to inflammation, which may serve as a significant risk factor. TME is also considered a potentially crucial mediator in this connection [[5], [6], [7]]. Analysing rectal cancer's TME, we studied SERPINA3 expression in epithelial and stromal cells and observed that epithelial SERPINA3 to be significantly elevated in tumors and metastasis, while stromal SERPINA3 was found higher in benign samples. This differential expression prompted an investigation into the association between SERPINA3 localization and rectal cancer progression. Earlier findings indicated that SERPINA3 staining was limited to tumor, normal, and stromal cells, including leucocytes and fibroblasts in CRC [20]. In contrast, immunostaining of colon tumor tissues demonstrated SERPINA3 expression in the cytoplasm and cell membrane, with no expression observed in normal and stromal tissues [21]. Another study showed predominant stromal expression of SERPINA3 in normal human endometrium [30], while cytoplasmic and extracellular expression found in endometrial epithelial cancer cells [31].
Notably, epithelial SERPINA3 expression exhibited a gradient from lower to higher levels during the progression from benign to metastatic tumors. Significantly elevated in moderately differentiated tumor grades and T ≥ 2 stage rectal adenocarcinomas, this finding aligns with a previous study indicating a positive correlation between SERPINA3 expression and metastatic potential in CRC [21]. Further, silencing SERPINA3 was shown to reduce invasion potential, linked to decreased levels of MMP2, MMP9, and a decrease in liver metastasis [21]. Overexpression of SERPINA3 in triple negative breast cancer (TNBC) cells resulted in significant upregulation of N-cadherin, vimentin, Snai1, Twist 1 and downregulated E-cadherin levels [32]. In CRC cells, SERPINA3 silencing up-regulated E-cadherin but down-regulated N-cadherin, Snail, and Vimentin [33]. Our study reveals heightened epithelial SERPINA3 expression in advanced-stage rectal adenocarcinomas, indicating its possible role in CRC progression. While the mechanism and downstream targets remain unclear, our findings suggest a critical role for epithelial SERPINA3 expression in cancer advancement. Interestingly, varying concentrations of treating intact SERPINA3 protein did not significantly affect the invasive potential of different CRC cell lines [20], leading us to speculate that the localization of SERPINA3, rather than just expression, may be a key determinant in CRC disease progression.
Our investigation revealed intriguing patterns of SERPINA3 expression in CRC, shedding light on its potential role in TME dynamics. Notably, stromal SERPINA3 exhibited higher expression in normal tissue, with a gradual decrease from benign to metastatic stages. This stromal association in early tumor formation suggests a critical role, warranting further exploration as prior studies have not extensively investigated SERPINA3's significance in CRC TME [22,23]. Our findings also demonstrated intense SERPINA3 staining in gastrointestinal stromal tumor and chronic inflammation samples (Fig. S2), emphasizing its potential involvement in pro-tumorigenic processes mediated by TASCs [34]. Interestingly, tumor stroma has a great influence on modulating the TME through immunosuppressive cells recruitment and prevention of effector immune cell infiltrations [35]. Studies have uncovered an upregulation of SERPINA3 in human placental disorders, correlating with hypomethylation of the 5′ (CpG island) promoter region of the gene [36]. This region potentially encompasses binding sites for proteins implicated in developmental processes and stress responses, including inflammation and hypoxia [37]. C-reactive protein, an inflammation marker showed a substantial positive correlation with plasma SERPINA3 in both CRC patients and controls [20]. Additionally, SERPINA3 has shown associations with inflammation across various diseases [[38], [39], [40]]. Given that the activation and regulation of SERPINA3 protein is predominantly influenced by inflammation response pathways, such as IL6, there is promising potential for further research into the relevance of SERPINA3 in the tumor microenvironment (TME). The IL-6-induced activation of the STAT3 pathway has been observed to enhance glycolysis in cells during chronic inflammation, potentially heightening their susceptibility to oncogenic transformation and maintenance [41]. Notably, in prostate cancer cells, circSERPINA3 has been identified as a stabilizer of SERPINA3, thereby inducing autophagy and aerobic glycolysis [42]. Considering STAT3's role as a direct transcriptional activator of SERPINA3, its involvement in inflammation-induced oncogenesis warrants further investigation. Our observations of elevated SERPINA3 protein levels in the stroma compared to the epithelium of inflammatory tissues versus their adenocarcinomas underscore the necessity for additional research to comprehensively grasp SERPINA3's role in the TME, especially within tumor-associated stromal cells. Subsequent investigations are imperative to explore differential tissue localization and elucidate the pro-tumorigenic functions of SERPINA3 in the TME.
SERPINA3 exhibits diverse immunostaining patterns in various cancers, being associated with poor prognosis. This variation is observed as predominant cytoplasmic staining in melanoma [43], lung carcinoma [44] and glioma [45], cytoplasmic and membrane staining in colon tumors [21], and cytoplasmic and extracellular localization in endometrial cancer cells [31]. In estrogen-stimulated breast cancer cells, SERPINA3 translocates from cytoplasm and nuclei to cytoplasmic granules [46]. Although some cancers display nuclear localization of SERPINA3 [44,47], intriguingly, high nuclear staining is correlated with better patient survival in liver cancer. Our study further identifies differential SERPINA3 localization in inflammatory tissues compared to adenocarcinomas, suggesting a subtle role in cancer progression. Notably, nuclear localization is prominent in higher stages or invading epithelia, while intact or lower-stage tumor epithelia exhibit predominantly cytoplasmic staining. Moreover, variations in SERPINA3's predominant localization are noted between cohorts, with nuclear expression in our Indian cohort and cytoplasmic staining in US BioMax tissue arrays. This discrepancy prompts a deeper exploration of the impact of inflammation on SERPINA3 localization and its potential compartment-specific functions.
Contrasting findings from a previous study indicate SERPINA3 in the nuclei of lymphoid cells invading stomach carcinoma masses, absent in rectal carcinoma mass nuclei, inflammatory areas like gastric ulcers or appendicitis, and normal tissues [48]. As the sole-nuclear binding secretory-serpin in the superfamily, SERPINA3 possesses DNA-binding sites despite lacking a specific nuclear localization signal [49]. Collectively, these studies underscore the necessity for comprehensive research to unravel the intricate roles of SERPINA3 in the CRC TME, encompassing its differential localization, downstream targets, and DNA binding dynamics [49].
In conclusion, our study identifies SERPINA3 as a promising marker for distinguishing EOCRC from late-onset cases. The differential expression patterns observed in both epithelial and stromal compartments suggest its potential role in the initiation and progression of EOCRC. These findings underscore the significance of SERPINA3 as a valuable tool for early diagnosis and understanding the distinct molecular factors associated with EOCRC. Further research is needed to deepen our insights in this area.
CRediT authorship contribution statement
Anjana Soman: Writing – original draft, Visualization, Methodology, Investigation, Conceptualization. Tapas Pradhan: Writing – review & editing, Visualization, Methodology, Conceptualization. R. Krishna: Validation, Investigation. Evangeline Surya Hermon: Validation, Investigation. Thara Somanathan: Formal analysis. Jinto Edakkalathoor George: Formal analysis. Gejoe George: Resources. Ramesh Pothuraju: Writing – review & editing, Visualization. S. Asha Nair: Supervision, Project administration, Funding acquisition.
Limitations of the study
We recognize the limitation of using a single pair of samples for RNA sequencing; however, the findings were subsequently validated through qRT-PCR and IHC, allowing us to draw meaningful inferences and conclusions. The study is constrained by the unavailability of a rectal cancer tissue array from the Indian cohort, and a need for a higher number of normal samples, and additional matched sample pairs for chronic inflammation and rectal cancer adenocarcinoma to enhance statistical significance.
Ethics statement
The study was approved by the institutional human ethics review committee (IHEC/1/2018/05). The patients/participants provided their written informed consent to participate in this study.
Data and code availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
This work was supported by the Department of Biotechnology (DBT), Government of India and research fellowship to AS from CSIR-UGC, Government of India.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Authors acknowledge the human subjects involved in the study. We thank Histopathology Department, RGCB for their immense help. We would like to thank cancer research program 4 (CRP4) laboratory members for the critical discussions and technical assistance. The figure was created by using BioRender software (https://www.biorender.com).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e40119.
Contributor Information
Ramesh Pothuraju, Email: rameshp@rgcb.res.in.
S. Asha Nair, Email: sasha@rgcb.res.in.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Cancer today. https://gco.iarc.who.int/today/en/dataviz/pie?mode=cancer&types=1&sort_by=value0&group_populations=1
- 2.Siegel Mph RL., Giaquinto A.N., Ahmedin |, Dvm J., Siegel R.L. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12–49. doi: 10.3322/CAAC.21820. [DOI] [PubMed] [Google Scholar]
- 3.Chou C.L., Chang S.C., Lin T.C., et al. Differences in clinicopathological characteristics of colorectal cancer between younger and elderly patients: an analysis of 322 patients from a single institution. Am. J. Surg. 2011;202(5):574–582. doi: 10.1016/j.amjsurg.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Cavestro G.M., Mannucci A., Zuppardo R.A., Di Leo M., Stoffel E., Tonon G. Early onset sporadic colorectal cancer: worrisome trends and oncogenic features. Dig. Liver Dis. 2018;50(6):521–532. doi: 10.1016/J.DLD.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad Kendong S.M., Raja Ali R.A., Nawawi K.N.M., Ahmad H.F., Mokhtar N.M. Gut dysbiosis and intestinal barrier dysfunction: potential explanation for early-onset colorectal cancer. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/FCIMB.2021.744606/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gausman V., Dornblaser D., Anand S., et al. Risk factors associated with early-onset colorectal cancer. Clin. Gastroenterol. Hepatol. 2019;0(0) doi: 10.1016/j.cgh.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutgens D., Vleggaar F.P., Schipper M.E.I.I., et al. High frequency of early colorectal cancer in inflammatory bowel disease. Gut. 2008;57(9):1246–1251. doi: 10.1136/gut.2007.143453. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton A.C., Bannon F.J., Dunne P.D., et al. Distinct molecular profiles of sporadic early-onset colorectal cancer: a population-based cohort and systematic review. Gastro Hep Adv. 2023;2(3):347–359. doi: 10.1016/j.gastha.2022.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawler T., Parlato L., Warren Andersen S. The histological and molecular characteristics of early-onset colorectal cancer: a systematic review and meta-analysis. Front. Oncol. 2024;14(April):1–13. doi: 10.3389/fonc.2024.1349572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballester V., Rashtak S., Boardman L. Clinical and molecular features of young-onset colorectal cancer. World J. Gastroenterol. 2016;22(5):1736–1744. doi: 10.3748/wjg.v22.i5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raman R., Kotapalli V., Adduri R., et al. Evidence for possible non-canonical pathway(s) driven early-onset colorectal cancer in India. Mol. Carcinog. 2014 doi: 10.1002/mc.21976. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo H., Betel D., Abelson J.S., Zheng X.E., Yantiss R., Shah M.A. Early-onset colorectal cancer is distinct from traditional colorectal cancer. Clin. Colorectal Cancer. 2017;16(4) doi: 10.1016/j.clcc.2017.06.002. 293-299.e6. [DOI] [PubMed] [Google Scholar]
- 13.Dozois E.J., Boardman L.A., Suwanthanma W., et al. Young-onset colorectal cancer in patients with no known genetic predisposition: can we increase early recognition and improve outcome? Medicine (Baltim.) 2008;87(5):259–263. doi: 10.1097/MD.0b013e3181881354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silla I.O., Rueda D., Rodriguez Y., Garcia J.L., Vigo F., Perea J. Early-onset colorectal cancer: a separate subset of colorectal cancer. World J. Gastroenterol. 2014;20(46) doi: 10.3748/WJG.V20.I46.17288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain S., Gautam V., Naseem S. Acute-phase proteins: as diagnostic tool. J. Pharm. BioAllied Sci. 2011;3(1):118. doi: 10.4103/0975-7406.76489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sánchez-Navarro A., González-Soria I., Caldiño-Bohn R., Bobadilla N.A. An integrative view of serpins in health and disease: the contribution of SerpinA3. Am J Physiol - Cell Physiol. 2021;320(4):C106–C118. doi: 10.1152/ajpcell.00366.2020. [DOI] [PubMed] [Google Scholar]
- 17.de Mezer M., Rogaliński J., Przewoźny S., et al. SERPINA3: stimulator or inhibitor of pathological changes. Biomed. 2023;11:156. doi: 10.3390/BIOMEDICINES11010156. 2023;11(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soman A., Asha Nair S. Unfolding the cascade of SERPINA3: inflammation to cancer. Biochim Biophys Acta - Rev Cancer. 2022;1877(5) doi: 10.1016/J.BBCAN.2022.188760. [DOI] [PubMed] [Google Scholar]
- 19.Cavallini A., Rotelli M.T., Lippolis C., et al. Human microRNA expression in sporadic and FAP-associated desmoid tumors and correlation with beta-catenin mutations. Oncotarget. 2017;8(26):41866–41875. doi: 10.18632/ONCOTARGET.16383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimberg J., Ström K., Löfgren S., Zar N., Hugander A., Matussek A. Expression of the serine protease inhibitor serpinA3 in human colorectal adenocarcinomas. Oncol. Lett. 2011;2(3):413–418. doi: 10.3892/ol.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao L.L., Pei X.F., Qiao X., et al. SERPINA3 silencing inhibits the migration, invasion, and liver metastasis of colon cancer cells. Dig. Dis. Sci. 2018;63(9):2309–2319. doi: 10.1007/s10620-018-5137-x. [DOI] [PubMed] [Google Scholar]
- 22.Yuan Q., Wang S.Q., Zhang G.T., et al. Highly expressed of SERPINA3 indicated poor prognosis and involved in immune suppression in glioma. Immunity, Inflamm Dis. 2021 doi: 10.1002/IID3.515. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Dong X., Cai J., et al. 2018. SERPINA3 Induced by Astroglia/Microglia Co - Culture Facilitates Glioblastoma Stem - like Cell Invasion; pp. 285–291. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedchenko N., Reifenrath J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue – a review. Diagn. Pathol. 2014;9:221. doi: 10.1186/S13000-014-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pradhan T., Kumar V., H E.S., et al. STIL endows oncogenic and stem- like attributes to colorectal cancer plausibly by shh and wnt signaling. 2021;11(August):1–15. doi: 10.3389/fonc.2021.581671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy C.C., Singal A.G. 2018. Establishing a Research Agenda for Early-Onset Colorectal Cancer. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paschke S., Jafarov S., Staib L., et al. Are colon and rectal cancer two different tumor entities? A proposal to abandon the term colorectal cancer. Int. J. Mol. Sci. 2018;19(9):1–24. doi: 10.3390/ijms19092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marx O., Mankarious M., Yochum G. Molecular genetics of early-onset colorectal cancer. World J. Biol. Chem. 2023;14(2):13–27. doi: 10.4331/wjbc.v14.i2.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terzi J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. YGAST. 138:2101-2114.e5. doi:10.1053/j.gastro.2010.01.058. [DOI] [PubMed]
- 30.Rj M., Sg B. Immunohistochemical demonstration of alpha-1-antitrypsin and alpha-1-antichymotrypsin in normal human endometrium. Int. J. Gynecol. Pathol. 1987;6(1):49–54. doi: 10.1097/00004347-198703000-00006. [DOI] [PubMed] [Google Scholar]
- 31.Yang G.D., Yang X.M., Lu H., et al. SERPINA3 promotes endometrial cancer cells growth by regulating G2/M cell cycle checkpoint and apoptosis. Int. J. Clin. Exp. Pathol. 2014;7(4):1348–1358. www.ijcep.com/ [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Tian J., Qu C., et al. Overexpression of SERPINA3 promotes tumor invasion and migration, epithelial-mesenchymal-transition in triple-negative breast cancer cells. Breast Cancer. 2021;1:3. doi: 10.1007/s12282-021-01221-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H., Wu X., Wang D., Li Q., Zhang X., Xu L. Unveiling the role of miR-137-3p/miR-296-5p/SERPINA3 signaling in colorectal cancer progression: integrative analysis of gene expression profiles and in vitro studies. BMC Med Genomics. 2023;16(1):1–16. doi: 10.1186/S12920-023-01763-W/FIGURES/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bussard K.M., Mutkus L., Stumpf K., Gomez-Manzano C., Marini F.C. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016;18(1):1–11. doi: 10.1186/s13058-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19(11):1423. doi: 10.1038/NM.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu S., Yin X., Zhang G., Meng F. Identification of DNA methylation signature to predict prognosis in gastric adenocarcinoma. J. Cell. Biochem. 2019;120(7):11708–11715. doi: 10.1002/JCB.28450. [DOI] [PubMed] [Google Scholar]
- 37.Chelbi S.T., Mondon F., Jammes H., et al. Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension. 2007;49(1):76–83. doi: 10.1161/01.HYP.0000250831.52876.cb. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto N.M., Aoki M., Okubo Y., et al. Gene expression profile of isolated dermal vascular endothelial cells in keloids. Front. Cell Dev. Biol. 2020;0:658. doi: 10.3389/FCELL.2020.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sergi D., Campbell F.M., Grant C., et al. SerpinA3N is a novel hypothalamic gene upregulated by a high-fat diet and leptin in mice 06 Biological Sciences 0604 Genetics. Genes Nutr. 2018;13(1) doi: 10.1186/S12263-018-0619-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinawath N., Vasoontara C., Jinawath A., et al. Oncoproteomic analysis reveals co-upregulation of RELA and STAT5 in carboplatin resistant ovarian carcinoma. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ando M., Uehara I., Kogure K., et al. Interleukin 6 enhances glycolysis through expression of the glycolytic enzymes hexokinase 2 and 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase-3. J. Nippon Med. Sch. 2010;77(2):97–105. doi: 10.1272/JNMS.77.97. [DOI] [PubMed] [Google Scholar]
- 42.Xing Z., Li S., Liu Z., Zhang C., Bai Z. CircSERPINA3 regulates SERPINA3-mediated apoptosis, autophagy and aerobic glycolysis of prostate cancer cells by competitively binding to MiR-653-5p and recruiting BUD13. J. Transl. Med. 2021;19(1):1–13. doi: 10.1186/S12967-021-03063-2/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J., Cheng Y., Tang L., et al. Up-regulation of SERPINA3 correlates with high mortality of melanoma patients and increased migration and invasion of cancer cells. Oncotarget. 2016;8(12):18712–18725. doi: 10.18632/ONCOTARGET.9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higashiyama M., Doi O., Yokouchi H., Kodama K., Nakamori S., Tateishi R. Alpha‐1‐antichymotrypsin expression in lung adenocarcinoma and its possible association with tumor progression. Cancer. 1995;76(8):1368–1376. doi: 10.1002/1097-0142(19951015)76:8<1368::AID-CNCR2820760812>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 45.Li Y., Dong X., Cai J., et al. SERPINA3 induced by astroglia/microglia co-culture facilitates glioblastoma stem-like cell invasion. Oncol. Lett. 2018;15(1):285. doi: 10.3892/OL.2017.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.I L., Ae L. Purification and characterization of an alpha 1-antichymotrypsin-like 66 kDa protein from the human breast cancer cell line, MCF-7. Biochim. Biophys. Acta. 1992;1121(1–2):119–129. doi: 10.1016/0167-4838(92)90345-E. [DOI] [PubMed] [Google Scholar]
- 47.Tahara E., Ito H., Taniyama K., Yokozaki H., Hata J. Alpha1-antitrypsin, alpha1-antichymotrypsin, and alpha2-macroglobulin in human gastric carcinomas: a retrospective immunohistochemical study. Hum. Pathol. 1984;15(10):957–964. doi: 10.1016/S0046-8177(84)80125-2. [DOI] [PubMed] [Google Scholar]
- 48.Takada S., Tsuda M., Mitomi T., et al. Analysis of the tissue and cellular localization of alpha-1-antichymotrypsin by an immunohistochemical technique. Gann, Japanese J Cancer Res. 1982;73(5):742–747. https://europepmc.org/article/med/6762318 [PubMed] [Google Scholar]
- 49.Naidoo N., Cooperman B.S., Wang Z mei, Liu X zhou, Rubin H. Identification of lysines within α1-antichymotrypsin important for DNA binding. AN unusual combination of DNA-BINDING elements. J. Biol. Chem. 1995;270(24):14548–14555. doi: 10.1074/JBC.270.24.14548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.