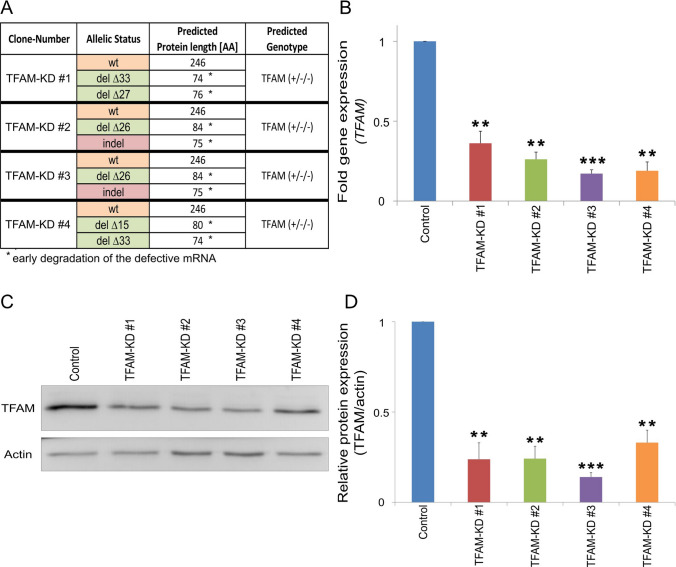
Fig. 1.
Validation of TFAM knockdown in colorectal cancer cells. After the introduction of deleterious mutations in TFAM using CRISPR-Cas9, four SW480 cell clones (TFAM-KD #1, TFAM-KD #2, TFAM-KD #3, TFAM-KD #4) were selected and TFAM mutations were confirmed through sequence analysis (A), changes in gene expression (B) and changes in protein expression (C, D). A Sequence analysis was used to identify the allelic status of cell clones and to predict the associated genotype. The TFAM protein length associated with this genotype was predicted with clustal omega. It should be noted that protein synthesis of mutated TFAM alleles is not expected to occur due to early degradation of defective mRNA. Sequencing of SW480 wild-type was used as a control (wt = wild-type, del = deletion, indel = insertion and deletion). B Gene expression of TFAM was measured by qPCR analysis and normalized to the reference gene GAPDH. Data from three experiments are shown and values represent means ± SEM (**p ≤ 0.01, ***p ≤ 0.001; paired t-test (two-tailed)). C, D Protein level of TFAM was measured and quantified by western blot analysis. 30 µg protein was loaded per lane and actin was used as a loading control. Values are means ± SEM (n = 3, **p ≤ 0.01, ***p ≤ 0.001; paired t-test (two-tailed))