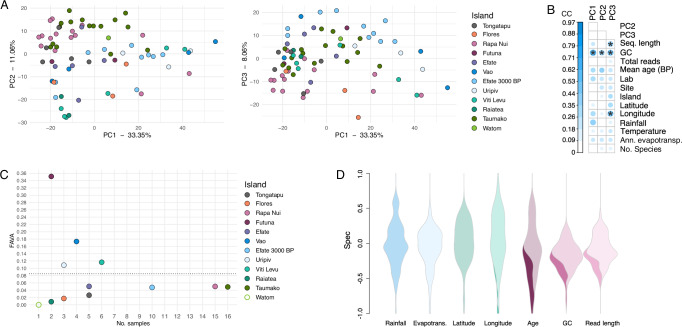
Fig. 2. Community species profiles are minimally structured by island.
A PCA of well-preserved calculus samples, coloured by the island from which samples were collected; PCs 1, 2, and 3 are shown, accounting for > 50% of the variation in the dataset. B Canonical correlation (CC) analysis comparing the positions of calculus samples in the PCA shown in (A) to environmental and laboratory metadata. Pearson two-sided correlation tests were used to determine if the correlations were significant. Metadata with a p ≤ 0.01 and CC value ≥ 0.4 are marked with an asterisk (*). Ann. evapotransp. - annual evapotranspiration; No. species - number of species. C FAVA values of variance in microbial composition between samples grouped by island. Dotted line indicates the FAVA value of all samples not separated by island. There is only one sample from Watom, so FAVA could not be calculated for this island, and the dot is left unfilled to indicate that FAVA is NA rather than 0. D Specificity of species in calculus samples to island conditions (blue), island location (green), or sequenced library characteristics (red). Dark colours in the violin plots indicate the proportion of species that are significantly associated with that metadata type. No dark colour indicates that no species were specifically associated with that metadata.