Abstract
Background
Many investigations have reported that advantageous mutations occurred more frequently under selective conditions than those under non-selective conditions. This phenomenon is referred to as adaptive mutation. Their characteristics are that adaptive mutations are directed and growth-independent. The idea of directed adaptive mutation had been objected by some reports, however, the idea of growth-independent adaptive mutation has been held till today.
Results
In this paper, we have observed that under leucine starvation conditions, leu+ revertants accumulated as a function of time; leu- to leu+ reverse mutation rates and frequencies were higher than those under non starvation conditions; and no divided cells could be monitored by the penicillin method. These results were similar to the time-dependent manner of adaptive mutation from previous reports. However, leucine concentration determinate experiments revealed that certain traces of leucine, which leaked from the E. coli cells, was almost always present in the culture. More numbers of leu+ revertants appeared when the similar cultures were dropped in small areas on the selective plates than when spread on the whole selective plates. These results have shown that mutations under leucine starving conditions are growth-dependent. Fluctuation analysis of leu+ revertants indicated that leu-leu+ mutation occurred spontaneously and randomly. In addition, the spectra of leuB gene in the revertants proved that mutations under selective conditions were not specific or directed.
Conclusions
The above investigations led to the conclusion (1) that the occurrence of leu+ mutations under starvation conditions was growth-dependent. The occurence mutations was also similar to that under non-starvation conditions (2). Under starvation conditions the mutation rates were higher, and was not constant during the long process.
Background
In 1988 John Cairns and co-workers [1] described an experimental system, in which bacteria, when plated under conditions where their growth was severely restricted by a single defective nutritional gene, mutated over several days to a phenotype that was able to grow. There are now many examples of this phenomenon. In some instances those late appearing colonies are actually slow growing mutants present in the culture at the time of plating. In many cases, however, the mutants can be shown to have arisen on the plate and it is characteristic that they do so at a rate higher than would be expected from the amount of genome DNA replication that occurs. This phenomenon has become known as "adaptive mutation", although other terms such as selection-induced mutation, stationary phase mutation, or starvation-associated mutation have also been employed. Nowadays, adaptive mutation is defined as a process that, during non-lethal selections, produces mutations that relieve the selective pressure whether or not the other, nonselective mutations are also produced [2].
During severe or prolonged selection, the adaptive mutations occurred at rates higher than those under non-selections. This observation is controversial to the New-Darwinism evolutionary theory: which assumed that the driving force of evolution is natural selection other than the creation of genetic variants, because the rate of mutation was presumably thought to be constant and unaffected by environments. The concept of mutation by Darwinism is different from that of Lamarckian. By Lamarckism, the adaptive mutations under selections were "directed", which means: (1) mutations arise among nonproliferating cells after selection is applied. (2) the presence of the selective agent is required for the mutations, (3) mutations that are not selected do not appear during selection. Although this idea of directed mutation had been rejected [3-6], another point of Lamarckism, which emphasized that the selective stresses had its roles on speeding up the mutation and evolution of organisms, was accepted by Darwin himself, and was supported by many experiments [7-14]. It was claimed that mutation rates under selective conditions were higher than those under non-selective conditions.
The widely used model bacterium for the study of adaptive mutation is Escherichia coil. Its growth restriction is usually realized by deprivation of a required amino acid or by supplying a sugar that cannot be metabolized. In this paper, we have chosen the leuB gene to investigate adaptive mutation for the following reasons: (1) the leuB gene has been sequenced in 1994 by Kirion et al [15]. (2) the reversion rate was conveniently low for using P0 method to calculate mutation rates [16]. (3) leucine is a substrate for cell growth, and is involved in a gene expression regulation system as an actor of leucine-responsive regulatory protein[17].
In this paper, the characteristics of reverse mutations and mutation rates during leucine starvation and non-starvation were compared. We found that the occurrence of mutations under starvation was growth-dependent, which was contrary to previous reports. Mutations under starvation occurred similarly to those under non-starvation, except at higher mutation rates. Based upon the experimental results we discussed the possible explanations about the role of selection on the mutation rates and evolution speed.
Results
Mutation rates increased during leucine starvation
On solid plates for leucine starvation, a reverse mutation formed only one leu+ revertant papilla on the flat colony. Therefore a papilla can be regard as one mutation event. The mutations accumulated in a time-dependent manner (Fig. 1). Leu+ revertant papillae did not appear until 2 days after plating due to the phenotype lag. The papillae observed in the first 2–3 days probably existed pre-plated. After that, new mutations continuously occurred and more papillae formed. These were regarded as mutations occurred under leucine starving pressure.
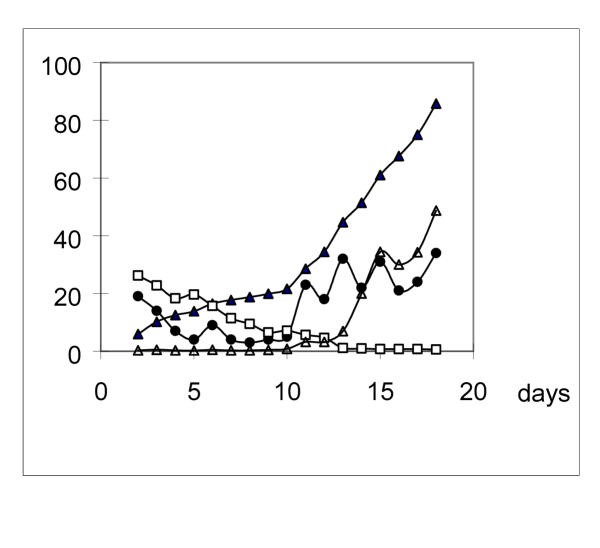
Figure 1.
Mutation rates increasing on leucine absent plates The rates were given as the number of leu+ revertants that appeared on a given day divided by the viable cells two days before. Solid triangle: Accumulation of leu+ revertants as papillae on colonies during leucine starvation. The data show the percent value of the ratio papillae per colony. Open square: Viable cells per colony with no papillae (107 cells/ml). Solid circle: Number of new mutations occurred during each day. Open triangle: mutation rates (10-8) in leucine starved colonies.
The results in Fig. 1 also indicated that reversion rates were not constant during the leucine starvation process. In the early period, fewer reversions took place per day; while in the later period, more reversions occurred per day. For example, during day 4–9, the average reversions per day were 5.14, but during day 14–18, the average reversions per day were 26.44. On the other hand, the number of viable cells decreased only about 17-fold during the same period. Thus the mutation rates were more than 100-fold higher during day 14–18 than that during day 4–9. This result was further supported by the data in Table 1 and Table 2. In Table 1, mutation rates of other types in liquid cultures also increased during starvation compared to those in rich medium-LB. In Table 2, especially on the micro-dropped plates, the number of revertant papillae per micro-drop increased with time of starvation.
Table 1.
Mutation rates and frequencies under different conditions in E. coli ATCC 31343
| Media | Time | Mutations | Viable cells | Mutation rates | Mutation frequencies |
| LB | 1 day | leu+ | 1.31 × 109 | 1.91 × 10-10 | 2.65 × 10-9 |
| LB | 1 day | pro+ | 1.24 × 109 | 1.41 × 10-10 | 6.57 × 10-9 |
| LB | 1 day | lac+ | 1.26 × 109 | 2.45 × 10-10 | 9.86 × 10-9 |
| MM+P | 1 day | leu+ | 7.56 × 107 | 5.73 × 10-9 | 3.02 × 10-7 |
| MM+L | 1 day | pro+ | 4.89 × 107 | 3.07 × 10-9 | 2.78 × 10-7 |
| CM(Lac) | 1 day | lac+ | 8.34 × 107 | 3.60 × 10-9 | 2.90 × 10-7 |
| MM+P | 3 days | leu+ | 2.13 × 107 | 1.72 × 10-8 | 1.11 × 10-6 |
| MM+L | 3 days | pro+ | 1.09 × 107 | 4.06 × 10-8 | 1.79 × 10-6 |
| CM(Lac) | 3 days | lac+ | 3.61 × 107 | 1.47 × 10-8 | 1.18 × 10-6 |
The initial number of cells in the cultures were all 1.08 × 108 cells/ml.
Table 2.
Leu+ reversions under selection were cell density-dependent
| Description | Number of micro-drops or plates with Ni papillae | ||||||
| Day 2 | Day 4 | Day 7 | Day 11 | Day 15 | Day 18 | ||
| A | 8.27 × 107 cells/micro-drop | N0 = 589 | N0 = 501 | N0 = 475 | N0 = 462 | N0 = 344 | N0 = 264 |
| Total micro-drops = 620 | N1 = 29 | N1 = 108 | N1 = 125 | N1 = 116 | N1 = 199 | N1 = 246 | |
| Total cells = 5.13 × 1010 | N2 = 2 | N2 = 11 | N2 = 13 | N2 = 20 | N2 = 49 | N2 = 63 | |
| N3 = 7 | N3 = 9 | N3 = 23 | N3 = 31 | ||||
| N4 = 3 | N4 = 5 | N4 = 13 | |||||
| N5 = 3 | |||||||
| Papillae/micro-drop | 0.053 | 0.210 | 0.277 | 0.315 | 0.619 | 0.858 | |
| B | 8.27 × 107 cells/plate | N1 = 6 | N1 = 8 N2 = 1 | N1 = 8 | N1 = 8 | N1 = 7 | N1 = 7 |
| Total plates = 50 | N2 = 2 | N2 = 2 | N2 = 2 | N2 = 2 | |||
| Total cells = 4.20 × 109 | N3 = 1 | N3 = 1 | |||||
| Papillae/plate | 0.12 | 0.20 | 0.24 | 0.24 | 0.28 | 0.28 | |
| C | 8.27 × 106 cells/micro-drop | N0 = 1600 | N0 = 1600 | N0 = 1600 | N0 = 1599 | N0 = 1598 | N0 = 1598 |
| Total micro-drops = 1600 | N1 = 1 | N1 = 2 | N1 = 2 | ||||
| Total cells = 1.32 × 1010 | |||||||
| Papillae/micro-drop | 0 | 0 | 0 | 0.001 | 0.001 | 0.001 | |
| D | 8.27 × 107 cells/micro-drop | N0 = 160 | N0 = 154 N1 = 6 | N0 = 152 | N0 = 145 | N0 = 137 | N0 = 127 |
| Total micro-drops = 160 | N1 = 8 | N1 = 15 | N1 = 23 | N1 = 33 | |||
| Total cells = 1.32 × 1010 | |||||||
| Papillae/micro-drop | 0 | 0.038 | 0.05 | 0.094 | 0.144 | 0.206 | |
| E | 2.96 × 108 cells/micro-drop | N0 = 45 | N0 = 41 | N0 = 33 | N0 = 21 | N0 = 11 | N0 = 8 |
| Total drops = 125 | N1 = 38 | N1 = 39 | N1 = 42 | N1 = 40 | N1 = 45 | N1 = 42 | |
| N2 = 24 | N2 = 24 | N2 = 27 | N2 = 37 | N2 = 40 | N2 = 38 | ||
| N3 = 11 | N3 = 12 | N3 = 13 | N3 = 12 | N3 = 13 | N3 = 19 | ||
| N4 = 6 | N4 = 7 | N4 = 8 | N4 = 12 | N4 = 11 | N4 = 12 | ||
| N5 = 1 | N5 = 2 | N5 = 2 | N5 = 3 | N5 = 5 | N5 = 6 | ||
| Papillae/micro-drop | 1.18 | 1.29 | 1.42 | 1.70 | 1.90 | 2.02 | |
| F | 2.96 × 108 cells/micro-drop | N0 = 125 | N0 = 125 | N0 = 125 | N0 = 125 | ------ | ------ |
| Total drops = 125 | |||||||
| Papillae/micro-drop | 0 | 0 | 0 | 0 | ----- | ------ | |
A,C,E: Cells were deposited as micro-drops on MM+P plates. B: Cells were spread on MM+P plates. D: cells were pre-cultured in 0.85% NaCl solution for 14 hours at 37°C before deposited as micro-drop on MM+P plates. F: Cells were deposited as micro-drops on MM+P plates with 100 μg/ml Ampcillin. Ampicillin was added once a day with 1 ml (1000 μg/ml) around the every micro-drops till day 7.
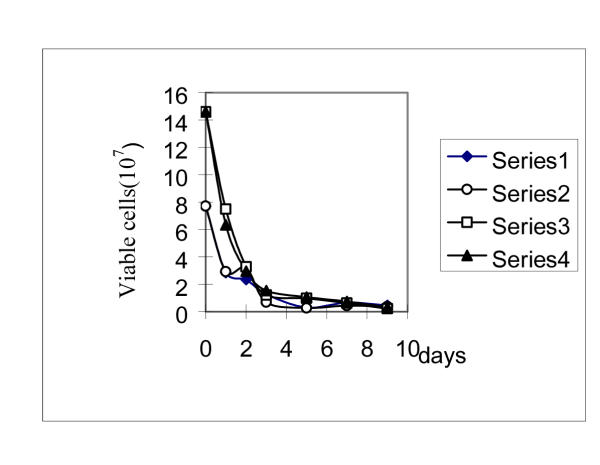
Death rate of cells on the spread selective plates was about 13.67% higher than that of on the dropped plates (Fig 2). But on both types of plate, the number of viable cells was similar regardless of the presence or absence of Ampicillin. It was previously regarded as the evidence that in the starvation process, cells did not divide, so adaptive mutations occurrence under selective conditions was regarded as cell division-independent [11], i.e., growth-independent. However the data in Table 2,3 were not in line with it.
Figure 2.
The viable cells on MM+P plates in with and without Ampicillin (100 μg/ml). Ampicillin was added onto the plates once a day by threading 1 ml Ampicillin solution (1000 μg/ml) around the whole plate surface. Each time, 3–5 micro-drops, which with no revertant papillae, were dug out and immerged into the tube with 1 milliliter 0.85% NaCl solution. After half an hour, vortex the tubes and diluted the suspension. The viable cells were counted by plating the appropriate aliquots onto LB plates after incubating at 37°C for 24 hours. Solid diamonds, viable cells in micro-drops on MM+P plate in the absence of Amp. Open circles, viable cells in micro-drops on MM+P plate in the presence of Amp. Solid triangles, viable cells in a whole MM+P plate in the absence of Amp. Open squares, viable cells in a whole MM+P plate in the presence of Amp.
Table 3.
Leucine leaked from scavenger bacteria supporting the cells' growth and mutation
| Number of cells per culture at starting time | Number of leu+ revertants in different media | |||||
| Time of scavenger bacteria culturing (day) | ||||||
| 0 | 6 hours | 24 hours | 48 hours | 120 hours | 168 hours | |
| 2.89 × 105 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.89 × 106 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2.89 × 107 | 0 | 0 | 0 | 1 | 2 | 1 |
| 2.89 × 108 | 0 | 1 | 1 | 5 | 4 | 5 |
| 2.89 × 109 | 0 | 2 | 3 | 17 | 21 | 17 |
| Total revertants | 0 | 3 | 4 | 23 | 27 | 25 |
| CFUA/culture | 2.31 × 105 | 1.36 × 106 | 3.54 × 106 | 8.53 × 106 | 3.73 × 106 | 3.02 × 106 |
| CFUB/culture | 7.64 × 107 | 1.69 × 108 | 3.23 × 108 | 3.69 × 108 | 4.46 × 108 | 4.10 × 108 |
| Leucine (ng/ml) | ------ | 578.7 | 842.3 | 971.9 | 963.2 | 962.1 |
A: The number of viable cells of 31343 which were cultured in the medium at 37°C for 24 hours. The initial number of 31343 cells were 2.89 × 105 cells/ml. The medium was MM+P in which the scavenger bacteria cells (in the dialysis tubings) of 35577 were pre-cultured for different days at the concentration of 7.97 × 108 cells/ml. B Same as A except that the initial number of 31343 cells was 2.89 × 108 cells/ml.
The leu+ reversions during leucine starvation were growth-independent
The leu+ reversions during leucine starvation were cell density-dependent
When same numbers of cells were spread on the whole MM+P plates or dropped in a limited small area on the MM+P plates for leucine starvation, similar amount of revertants could be seen in the first several days, but in the following days, fewer revertants appeared on the spread plates than those on the dropped plates, and the revertants accumulated as a function of time on the dropped plates (Table 2 and Fig. 3).
Figure 3.
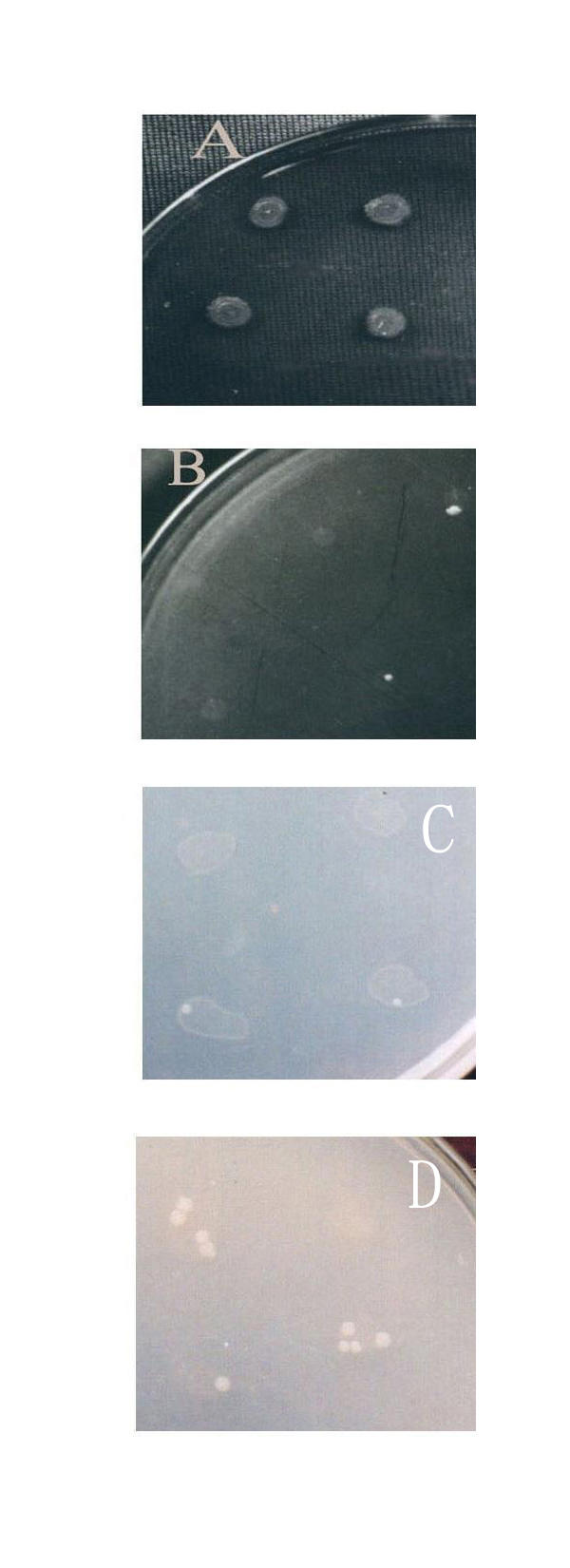
Leu+ revertants accumulating in the micro-drops as a function of time. A: On CM plate(day 12), without papillae can be seen on the thicker mosses. B: On MM+P plate(day 5), papillae can be seen on the thin mosses. 1.87 × 107 cells per drop. C: On MM+P plate(day 5), papillae can be seen on the thin mosses. 2.96 × 108 cells per drop. D: On MM+P plate(day 12), papillae can be seen on the thin mosses. 2.96 × 108 cells per drop.
In Table 2, the results showed that in dropped plates (Experiment A, E), all the cells were heaped in a small area and a small proportion of cells could divide by utilizing enough leucine leaked from the other viable or dead cells, then the reverse mutations could occur continuously in a time-dependent manner. However, when the same numbers of cells were spread on a whole plate, (the area of a plate was about 400 times larger than the area of a micro-drop), the cells were so dispersed that they could not absorb enough leucine to support them to grow and divide, and the reversion rate was much lower than those on the dropped plates, especially in the later culturing period (Experiment B). When the same numbers of cells were incubated in 0.85% NaCl solution overnight at 37°C for exhausting the nutrients in vivo of the cells before they were dropped on the plates, much fewer revertants were resulted (Experiment D). When the same numbers of cells were dropped on the MM+P plates (which contains 100 μg/ml Ampicillin), no revertants were found (Experiment F). If fewer cells were dropped on the plates, almost no revertants could be seen (Experiment C). All these results suggested that the reverse mutation under leucine starvation was cell density dependent and growth-dependent. And this conclusion was further supported by the following results in Table 3,4 and Fig 4.
Table 4.
The percentage of leucine to the total protein in vivo of 31343 cellsA
| MediaB | Leu+ revertant | Leu- revertant | ||||
| Leu% | Ala% | Lys% | Leu% | Ala% | Lys% | |
| LB | 9.074 | 9.796 | 7.275 | 8.840 | 9.792 | 7.260 |
| CM | 8.713 | 9.640 | 7.804 | 7.738 | 9.710 | 7.477 |
A The data is the mean of three experiments. B: The cells were cultured in the media LB and CM over night before they were collected and analyzed.
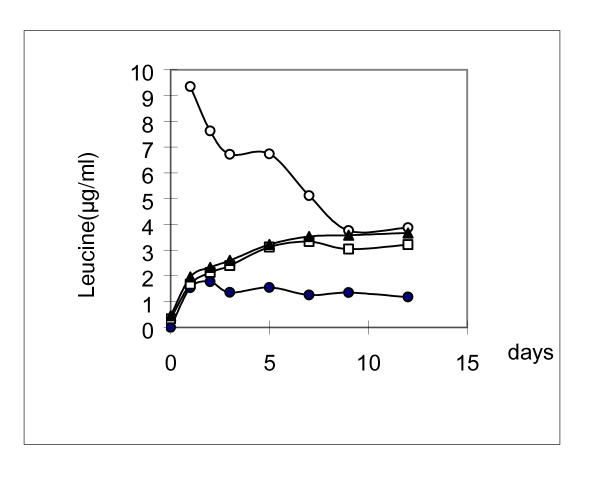
Figure 4.
The concentration of leucine in different culture media containing 1.8 × 109 cells/ml. The cells were pre-cultured in LB medium. Solid circle, in 0.85%NaCl solution; Open square, in MM medium; Solid triangle, in MM+P medium; Open circle, in CM medium
Leucine leaked from the scavenger cells supporting strain ATCC31343 cells' growth and mutation
When the scavenger bacterium cells of strain ATCC35577 (in dialysis tubings) were transferred into the MM+P medium, leucine was found to diffuse into the medium in several hours and approached to the highest point at day 2 and maintained at a stable level. The leu+ reversions of cells of strain ATCC31343 increased when the leucine concentration increased (Table 3). This result provided another evidence that the occurrence of leu+ mutation was growth-dependent.
The concentration of leucine in the cells and in the media
The concentration of leucine in the cells and in the media was determined using HITACHI 835 amino acid analyzer. The percentage of leucine to the total amount of 19 amino acids (tryptophan excepted) in vivo of strain ATCC31343 cells was summarized in Table 4. It could be seen that the percentage of leucine from cells inoculated in rich medium (LB) was higher than that of cells inoculated in synthesized complete medium (CM). Once the cells gathered from LB or CM were added into the 0.85% NaCl solution, the leucine quickly diffused from the cells into the solution. After about 6 hours in NaCl solution, the level of leaked leucine approached the highest point and sustained for a long time (Fig. 4). In MM and MM+P media, in which leucine was absent, the amount of leucine was similar to that in NaCl solution. In CM medium, the amount of leucine dramatically decreased in the first few hours, and then the decreasing rate slowed down and maintained at a stable level of a little higher than that of in MM and MM+P. These results suggested that the amount of leucine leaked into the culture was sufficient for the growth and division of a few cells during the prolonged incubation for leucine starvation. The number of divided cells was dependent upon the total numbers of the cells in the medium at the start time and the type of medium in which the cells were inoculated. When there was no leucine present, there would not be any cell growth or mutation.
Mutations under selective conditions were spontaneous
Fluctuation analysis of leu-leu+ mutation
According to the hypothesis of adaptive mutation by Cairns et al, the distribution of mutations under selection should follow Poisson pattern [1]. But the statistical data in Table 5 revealed that under different selective conditions, there were significant difference between the experimental data and the predicted data by Poisson distribution. This agreed very well with the general principles of spontaneous and random mutations in bacteria [16].
Table 5.
Fluctuation tests and analysis of mutations under selective conditions
| Exp.>A | Media /Mutation | Cultures with no mutants/total cultures | Number of mutants | MeanB | VarianceC | Distribu-tionD |
| 1–1 | LB leu-leu+ | 3/10 | 1,1,1,2,2,2,2 | 1.10 | 0.767 | P |
| 1–2 | CM leu-leu+ | 0/10 | 16,16,18,19,21,21,24, 24,27,28 | 22.60 | 18.267 | P |
| 1–3 | MM+P leu-leu+ | 0/10 | 519,552,526,612,572,634,632,619, 601,597 | 586.4 | 1774.5 | P |
| 1–4 | PB leu-leu+ | 0/10 | 1,1,2,2,2,,3,3,4,5,8 | 3.10 | 4.545 | P |
| 2–1 | LB leu-leu+ | 34/45 | 3,16,27,37,85,116,146,326,680,372,1284 | 90.64 | 63989 | LD |
| 2–2 | CM leu-leu+ | 16/45 | 2,5,7,7,9,9,10,12,13,14,14,16,16,16,18,18,19,19,30,35,42,44, 48,50,62,75,104,126,231 | 23.80 | 1770.3 | LD |
| 2–3 | MM+P leu-leu+ | 22/45 | 1,2,2,3,3,4,6,6,8,8,9,10,11,13, 15,19,23,24,25,32,55,67,103 | 9.98 | 402.20 | LD |
| 3–1 | MM+P leu-leu+ | 14/45 | 1,1,1,3,3,4,4,13,21,66 | 4.875 | 193.42 | LD |
| 3–2 | MM+P leu-leu+ | 4/24 | 1,1,1,2,2,3,3,3,4,4,5,5,7,7,9,12,12,19,26,50 | 7.333 | 123.19 | LD |
| 3–3 | MM+P leu-leu+ | 11/24 | 1,1,1,2.3.7.9.12.14,21,29,33,38 | 7.125 | 134.03 | LD |
| 4–1 | MM+L pro-pro+ | 14/24 | 1,1,1,1,3,4,9,13,15,41 | 3.708 | 80.65 | LD |
| 4–2 | MM+L Pro-pro+ | 14/24 | 1,1,1,2,2,2,4,5,7,9,18,34 | 3.583 | 59.04 | LD |
| 4–3 | MM+L pro-pro+ | 14/24 | 1,1,1,1,2,2,3,3,5,11,11,12, 24,42 | 4.958 | 95.26 | LD |
| 5–1 | CM-lac lac-lac+ | 19/24 | 1,1, 4,23,38 | 2.792 | 78.43 | LD |
| 5–2 | CM-lac lac-lac+ | 14/24 | 1,1,1,1,2,2,2,6,30,44 | 3.750 | 110.89 | LD |
| 5–3 | CM-lac lac-lac+ | 11/14 | 1,1,1,2,2,2,3,3,5,6,9,14,72 | 5.042 | 214.99 | LD |
A: Experiments in group 1 (1–1 to 1–4) , aliquots from cultures in same flasks; experiments in other groups, each cultures were independent in a little tube or in a well of 48 wells microtittle. B: Mean of mutants. ![]() , here, n = number of cultures, xi = number of mutants in a culture. C: Variance of simples.
, here, n = number of cultures, xi = number of mutants in a culture. C: Variance of simples. ![]() D: distribution patterns were judged by the Value of X2.
D: distribution patterns were judged by the Value of X2. ![]() n = 10□ a0.95 = 3.94; n = 24, a0.95= = 13.848; n = 45 a0.95 = 30.612. If the value of X2 > a0.95, the distribution type was non-Poisson pattern; If the value of X2 < a0.95, the distribution type was Poisson pattern.
n = 10□ a0.95 = 3.94; n = 24, a0.95= = 13.848; n = 45 a0.95 = 30.612. If the value of X2 > a0.95, the distribution type was non-Poisson pattern; If the value of X2 < a0.95, the distribution type was Poisson pattern.
Sequence analysis of leuB gene of revertants
The sequence of the leuB gene, which encodes the enzyme β-isopropyl malate dehydrogenease (EC 1.1.1.85), has been determined [15] (GenBank accession no. D17631). Sequence analysis of the leuB gene from the leuB- mutant ATCC31343 revealed complete nucleotide sequence identity to the published leuB gene except for a C-to-T transition at nucleotide 857. This point mutation would result in a serine-to-leucine substitution at amino acid residue 286 of the LeuB protein. The sequence of the leuB+ from various revertants in the region of the mutation was shown in Table 6 and the possible single base substitutions in this cordon and the corresponding amino acids were also summarized.
Table 6.
Sequence of the leuB- mutant and leuB+ revertants
| Signal base substitution Sequence predicted at 856–858 | Amino acid residue 286 | Description |
| TTG | Leucine | LeuB- mutant |
| TCG | Serine | Wild type |
| GTG | Valine | Phenotypic wild type |
| ATG | Methionine | Phenotypic wild type |
| TTG | Leucine | Suppressor mutant |
| Amino acid residues in revertants | Description | |
| Amino acid 286 | Strain Type (Number) | |
| Ser | X(4), Y(2), Z(1) | X: Isolated from solid Leucine Starvation plates |
| Ser | X(2), Y(3), Z(2) | Y: Isolated from liquid Leucine Starvation cultures |
| Met | X(2), Y(4), Z(1) | Z: Isolated from non-Starvation plates and cultures |
| Leu | X(3), Y(2), Z(1) |
According to the report by Wright et al [18], there are three general classes of leuB+ revertants: (1) true revertants, with base substitutions encoding serine (wild type), (2) revertants with base substitutions encoding valine and methionine, these revertants had the growth rates and generation time similar to the wild type. (3) suppressors, which still retained the Leu-286 residue of the parental leuB mutant, but usually had lower growth rates and longer generation time than the wild type.
The spectra of leuB gene of 22 revertants (Table 6), which were isolated under leucine starvation conditions (X+Y), included all the three types of reverse mutations as reported by Wright et al [18]. Their ratio of the three types is: Ser: Val: Met = 16:18:2. Our data is Ser: Val: Met = 6:5:6. This suggested that the occurrence of leu+ mutations under leucine starvation have no specificity. It was in accordance with the report by Hall [6], but not with the other reports [19].
Discussion
Mutations occurrence under leucine starvation were growth-dependent
The data in Table 2 and Table 3 indicated that the occurrence of leu+ reverse mutations was growth-dependent under leucine starvation conditions. Although the death rates of cells on the spread MM+P plates and on the dropped MM+P plates was similar with or without the addition of Ampicillin (Fig. 2), and the reverse mutations under leucine starvation increased with the time prolonged (Fig. 1). These two results were similar to the previous investigations reported by Hall [9]. They were previously regarded as the evidence that in selective conditions, cells did not divide, and adaptive mutation was growth independent [11]. Our explanation for this discrepancy is that: under selective conditions, only small a part of cells divided, and these divided cells were so few compared to the mass of the viable but non-dividing cells that they could not be monitored by the penicillin method.
Although the cells in divisions may only account for a small proportion of the total viable cells under leucine starving conditions, they might become the important source for the appearance of leu+ revertants. This was consistent with the results from Table 2, which indicated that the occurrence of mutation was cell density and growth dependent, and was further supported by the results in Fig. 4 of the leucine concentration determination and by the reversions occurred in the media where scavenger bacteria were pre-cultured (Table 3). Besides, extracellular DNA and nucleotides could also be consumed as the source of carbon and energy supporting the bacteria cells to grow under nutrient deprived conditions [20]. This also suggested that a small proportion of the population cells could grow in nutrient deprived conditions by absorbing the amino acids and peptides or absorbing DNA and nucleotides, which may leak from the other viable or dead cells.
It has already been proved that mutations under selection require trace nutrient which the cell is defect in the function to synthesize or metabolize. Hall used minimal medium containing 5 μM tryptophan to select trp+ revertants [9], Fani [7] reported the number of his+ revertants was always greater in plates containing trace of histidine (0.25 ug/ml) than those without histidine in E. coli FB184 hisA. All these results also suggested that occurrence of mutations under selection were growth-dependent.
Mutations occurred spontaneously under selections
Fluctuation analysis of mutations in liquid culture indicated that mutations occurred spontaneously during leucine starvation. It is well known that mutations occurred spontaneously under nonselective conditions in bacteria. From the results 2.3, we could see that under starving condition, there was still trace amount of leucine leaked from other cells in the medium, the reverse mutations could originated from the minority divided cells by the support of this amount of leucine, and then would have the Luria-Delbrück distribution patterns.
The spectra of leuB gene of leu+ revertants, isolated from leucine starvation conditions, did not show specificity. This was in agreement with our conclusion that mutations under starvation were growth dependent and occurred spontaneously.
Mutation rates were not constant during leucine starvation
Our results in fig. 1 indicated reversion rates under leucine starvation condition were not constant. In the first few days (2–3 days, in Fig. 1), the leu+ reversions accumulated quickly, and they were regarded as existing pre-plated and formed papillae at different time intervals because of the phenotype lag. In the days followed, new reversions accumulated very slowly, and the mutation rate during this period (day 4–10) was low. Later on, reversions accumulated more quickly, and the mutation rate was high. This stage could sustain for some time (day 12–21) and decreased at last.
Although other researchers have reported the mutation rates increased under selective conditions, they did not mention or emphasis that the mutation rates were not constant. For examples, Hall [9,10] used E. coli FCY2B trpB- mutant, E. coli FCY6B trpAB- mutant, on tryptophan limited plates; Shapiro[14] used E. coli MCS2 ara-lacZ, on Ara-Lac selection plates with different concentrations of glucose; Bridges[21] used E. coli trp A23 mut Y under on minimal plates; Kasak[22] used P. Putica PaW85 phe- on phenol-minimal plates. All of curves (the mutants appeared in a day versus culturing time) involved several parts which with different curvatures respectively. They could not be simply regarded linear as Hall did [9].
In fact, during the selection process, the viable cells did not increase but decreased slowly. This is an important reason for elevating the mutation rate. But, the magnitude of viable cells decreased was less than that of increased mutation rate, which suggested the selection played a role on elevating the mutation rates.
Why were mutation rates inconstant during selections, and were higher under selections than that of under non-selections? Here are some possible explanations.
Firstly, there was misapprehension for defining and calculating mutation rate. Generally, mutation rate (μ) was defined as mutations per cell per division. For a certain gene in a bacterium such as E. coli, regardless of the hot spots, suppose that the generation time is about constant during the exponential phase and all the cells are synchronized, μ is proportional to the mutation rate μb (mutations per base pair per genome replication), according to Drake's paper [23]. For bacteria growing in rich medium (such as LB) in the exponential phase, the number of cells is approximate to the number of dividing cells. So, in the normally growing populations in exponential phase, almost each originating of DNA replication can result in a complete genome and accompany a cell division, μb is thus almost equals to the number of damages or mistakes per base pair occurs during one genome replication, and μb is low.
Under selections, the divided cells are so few that it is difficult to determine the number of divided cells. Then the mutation rate is usually defined as mutations per viable cell per day[7,9,10,12,14], marked as μd in this paper. If the generation time is constant during the selective process, μd is also proportional to the mutation rate μ and to μb .
Under selections, the growth of E. coli cells was severely restricted. The generation time is much longer than that under non-selective conditions. During the prolonged period of cell division, although most cells cannot replicate complete genomes, they might have initiated genome replications and produced incomplete genomes. This assumption was based on the result that 25% of the genome turned over per day during amino acid starvation as estimated by Bridges from the mutation rate of a mutT mutant [24], and based on that 20% of genome DNA was turned over in 24 hours in the buffer determined by Tang [25]. Of the whole population, where small portion of cells accomplish complete genome replication, most of cells would undertake incomplete replication. Thus the accumulated spontaneous DNA damages in the uncompleted cycles, which would cause mutations when DNA replication resumes, may be fixed in the completed genome, and become true mutations. Different pathways may be used to fix DNA damages and an important one is homogenous recombination. The long generation time provides opportunities for cells to fix these DNA damages. So the mutations per base per genome replication not only include the damages in one cycle of complete genome DNA replication, but also include any accumulated DNA damages in other incomplete genome DNA replications. Thus, under selective conditions, mutation rate per base pair per genome replication becomes a collective result and would be higher than that under non-selection. Due to the same reason, a bacteria population at different growth stages during the selection progress may have different generation times and would result in different mutation rates respectively.
Secondly, changes of metabolic control system under selective conditions may also elevate the background mutation rates, which can also affect advantageous adaptive mutation rates. It is well known that in the long process of evolution of microbes, diverse mechanisms had been developed to maintain their genetic stability (μg) and decrease their mutation rates (μb) during their genome expanding[23]. These mechanisms may include metabolic controls over concentrations of endogenous and exogenous mutagens, pre-replication DNA repair systems, the insertion accuracy of polymerases, 3'-exonucleolytic proofreading, and several post-replication systems for repairing mismatches [2,26-36]. Different bacteria apply different sets of these mechanisms, and the efficiency of a particular mechanism varies among microbes.
Leucine starvation and other selective condition can be regarded as an environmental signal. Populations that have experienced a severe challenge from the selective environment might therefore be expected to change their physiological state extensively, such as stringent response, global regulation in gene expressions, and other inducible responses such as SOS. These changes are to keep cells surviving and adapting to the selective environment. During this process, the elaborate enzymatic machinery, which can keep the fidelity of DNA replication and control the spontaneous mutation rate, are destroyed; some new proteins must be synthesized using exogenous and endogenous nutrition and energy sources. In the end, populated cells could get into a relative stable states, in which, cell division becomes more difficult, generation time becomes longer; the fidelity of DNA replication decreases, the spontaneous mutation rates (adaptive mutation rates) increases.
Conclusion
From the above discussions, we reached the conclusion that mutations under leucine starvation were growth-dependent. Mutations can occur spontaneously either under starvation and non-starvation; and show no specificity in the sequences of leuB genes between the leu+ revertants isolated from leucine starvation. The higher mutation rate under starvation could be caused by two reasons. One is an error in defining and calculating mutation rate; the other by the changes of metabolic control system under selective condition, which can elevate the background mutation rates, including the advantageous mutations.
Our explanation for the higher mutation rates under selective condition is not an inconsistency with other mechanisms of adaptive mutation, such as "hupermutable state" model [9,29,30]; homogenous recombination mechanism [2,14,31,32]; and the functions of mismatch repair system and mutator alleles [31-37]. The results could provide valuable information for further understanding of the relationship between environmental conditions and speed of evolution.
Materials and Methods
Strains
The strain used in this study was purchased from ATCC.
Escherichia coli ATCC 31343 F- hsdS20 ((r- B m- B)) ara-14 leuB6 proA2 lacY1 galK2 rpsI20 xyl-5 mtl-1 supE44 lambda-.
Escherichia coli ATCC 35577 F- recA1 uvrA6 phr-1 thi-1 thr-1 leuB6 lacY1 galK2 ara14 xyl15 mtl1 proA2 argE3 rpsL31 tsx-33 supE44 gyrA98 lambda-.
Media
The rich complete medium was Luria broth (LB), consisting of 10 g of peptone, 5 g of yeast extract powder, and 10 g NaCl per liter. pH7.2. Minimal medium (MM) consisting of 0.5 g of sodium citrate, 7 g of K2HPO4·2H2O, 3 g of KH2PO4·2H2O, 1 g of (NH4)2SO4, 0.1 g of MgSO4·7H2O, 2 g of glucose, and 1 mg of thiamine per liter. Selective medium containing, in addition to the minimal medium, 30 mg leucine per liter for selecting pro+ mutants (MM+L), or 30 mg proline for selecting leu+ mutants (MM+P). Complete synthesized medium (CM) containing, in addition to minimal medium, 30 mg leucine and 30 mg proline per liter. CM-Lac medium was used to select lac+ revertants, it was same to CM except that glucose was replaced by lactose.
Growth conditions for mutation rate and mutation frequency determinations on solid plates
Cells were grown at 37°C for 12–16 hours in LB medium. Then the cells were centrifuged, washed, and resuspended to about 1 × 109 cells/ml. The 10 μl aliquots were distributed on the MM+P selective plates and on CM plates. All plates were incubated in a humidified chamber at 37°C. On dropped plates, the suspension formed "flat colony" that was normal in appearance except for a slight dimpled. The number of leu+ revertants papillae on the flat colonies was counted each day; and the number of viable cells in the flat colony was determined by transferring them onto LB plates.
On solid plates, only one papilla formed after a mutation occurred. So, the number of mutations is approximately equal to the number of revertant papillae. The mutation rates is determined according to the expression μ = M/N, where M is number of reverse mutations, is about equal to the number of revertant papillae Nm. N is the number of viable cells in the flat colonies with no papillae, these colonies were present on the plates 2 days earlier than the papillae counted. The mutation frequency is determined according to the expression f = Nm/N.
Growth conditions for mutation rate and mutation frequency determinations in liquid cultures
Cells were grown at 37°C for 12–16 hours in LB medium . Then the cells were centrifuged, washed, and resuspended to about 1 × 109 cells/ml. The 2-ml aliquots were transferred into 18 ml MM+P, CM and LB medium, respectively. A serial of 0.5 ml aliquots from each well mixed suspension were dropped into 48 wells of a microtilter dish and incubated at 37°C. The number of leu+ revertant were determined by plating the cultures onto MM+P plates, the number of viable cells were determined by diluting and plating the cultures onto LB plates.
The mutation rates were calculated by the P0 methods of Luria and Delbrück [16]. According to the equation of μ = -lnP0/N, where P0 is the proportion of cultures with no revertants, and N is the mean number of cells per culture. The mutation frequency is determined according to the equation of f = ∑Nm/nN. Where, ∑Nm is the total number of revertants in all cultures, N is the mean number of cells per culture, and n is the number of cultures.
PCR amplification and sequencing the leuB gene
The PCR primers were based upon the nucleotides sequence from the GenBank (accession no.D17631) The primer 1 is 5'-gctaccgatattgtcgagtc-3', 110 bp upstream the initiate code 'atg'; the primer 2 is 5'-cgtcttagccatgattacac-3', 14 bp downstream the terminate code 'taa'. The PCR product is 1216 bp, containing the full length of leuB gene (1092 bp).
All PCR reactions and PCR products purification have done according to the manufacturer's instructions of Qiagen PCR kit. The purified PCR products were separated and sequenced by cycle sequencing according to the manufacturer's instructions on an ABI Model 3700 automated sequencer by United Gene Holdings, Ltd.(Shanghai, China).
Determination of the leucine concentrations during the process of cells' growth
Cells were grown at 37°C for 12 hours in LB medium. Then the cells were centrifuged, washed, and resuspended. The aliquots were distributed in 0.85%NaCl, MM, MM+P and CM media respectively, and continuously incubated at 37°C. After every period, part of the cultures were picked off to determination the number of viable cells by transferring appropriate culture onto LB plates. The leucine concentrations in the cultures (discarding the cells) and in the supernatants of the cells lysates were determined by using HITACHI 835 high speed amino acids analyzer. See additional file 1, 2 for the information about this analyzer and the determination conditions (Additional File 1 and Additional File 2).
Authors' Contributions
Ph.D Jianling Jin carried out all the molecular genetics studies except the sequencing of leuB genes of revertants, and drafted the manuscript. Peiji Gao, as supervisor, conceived the study, participated in its design and revised the manuscript. Yumin Mao carried out the sequencing of the leuB genes of revertants.
Supplementary Material
The information about HITACHI 835 high speed amino acids analyzer.
The leucine concentration determination conditions.
Acknowledgments
Acknowledgements
We thank Da Liu and Jianhua Zhang for their help in some experiments. Thank Ph.D. Weifeng Liu and Ph.D. Ke Gao for revising this manuscript. We also thank PL Forster, JA Shapiro, BA Bridges and SM Rosenberg for delivering their published papers to us. This work was supported by grant 39997004 from the Chinese National Science Foundation.
Contributor Information
Jianling Jin, Email: jjl@life.sdu.edu.cn.
Peiji Gao, Email: gaopj@sdu.edu.cn.
Yumin Mao, Email: ymmao@fudan.edu.cn.
References
- Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Foster PL. Adaptive mutation: implications for evolution. Bioassays. 2000;22:1067–1074. doi: 10.1002/1521-1878(200012)22:12<1067::AID-BIES4>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JE, Lenski RE. New data on excisions of Mu from E. coli MCS2 cast doubt on directed mutation hypothesis. Nature. 1990;344:173–175. doi: 10.1038/344173a0. [DOI] [PubMed] [Google Scholar]
- Miller JE, Lenski RE. Experimental evidence for an alternative to directed mutation in the bgl operon. Nature. 1992;356:446–448. doi: 10.1038/356446a0. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD. A test of the directed mutation hypothesis in Escherichia coli MCS2 using replaca plating. J Bacteriol. 1995;177:1119–1120. doi: 10.1128/jb.177.4.1119-1120.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Spectra of spontaneous growth-dependent and adaptive mutations at ebgR. J Bacteriol. 1999;181:149–155. doi: 10.1128/jb.181.4.1149-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani R, Callo R, Fancelli S, Mori E, Tamburini E, Lazcano A. Heterologous gene expression in an E. coli population under starvation stress conditions. J Mol Evol. 1998;47:363–368. doi: 10.1007/pl00013149. [DOI] [PubMed] [Google Scholar]
- Foster PL, Cairns J. The occurrence of heritable Mu excisions in starving cells of Escherichia coli. EMBO J. 1994;13:5240–5244. doi: 10.1002/j.1460-2075.1994.tb06855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Adaptive evolution that requires multiple spontaneous mutations: Mutations involving base substitutions. Proc Natl Acad Sci USA. 1991;88:5882–5886. doi: 10.1073/pnas.88.13.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Adaptive mutation: the uses of adversity. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.micro.47.1.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizatullin FS, Babynin EV. The selection induced His+ reversion in Salmonella typhimurium. Mutat Res. 1996;357:43–56. doi: 10.1016/0027-5107(96)00078-4. [DOI] [PubMed] [Google Scholar]
- Hall BG. Selection-induced mutations occur in yeast. Proc Natl Acad Sci USA. 1992;89:4300–4303. doi: 10.1073/pnas.89.10.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel G, Shapiro JA. The roles of starvation and selection in the emergence of araB-lacZ fusion clones. EMBO J. 1994;13:5229–5244. doi: 10.1002/j.1460-2075.1994.tb06854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirion H, Acki M, Aoshima M, Hayashi Y, Ohba M, Yamagishi A, Wakagi T, Oshima T. Hydrophobic interaction at the subunit interface contributes to the thermostability of 3'-isopropylmalate dehydrogenase from an extreme thermophile, Thermus thermophilus. J Bacteiol. 1994;144:499–508. doi: 10.1111/j.1432-1033.1994.tb18623.x. [DOI] [PubMed] [Google Scholar]
- Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo JM, Methews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE, Minnick MF. Reversion rates in a leuB autotrophy of E. coli K-12 correlate with ppGpp levels during exponential growth. Microbiology. 1997;143:847–854. doi: 10.1099/00221287-143-3-847. [DOI] [PubMed] [Google Scholar]
- Hall BG. Spectrum of mutations that occur under selective and non-selective conditions in E. coli. Genetica. 1991;84:73–6. doi: 10.1007/BF00116545. [DOI] [PubMed] [Google Scholar]
- inkel SE, Kolter R. DNA as a nutrient: novel role for bacteria competence gene homologes. J Bacteriol. 2001;183:6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges BA. Elevated mutation rate in mutY bacteria during starvation: evidence for DNA turnover. J Bacteriol. 1996;178:2709–11. doi: 10.1128/jb.178.9.2709-2711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasak L, Horak R, Kivisaar M. Promoter-creating mutations in Pseudomonas putida: A model system for the study of mutation in starving bacteria. Proc Natl Acad Sci USA. 1997;94:3134–3139. doi: 10.1073/pnas.94.7.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges BA, Ereira S. DNA sythesis and viability of a mut T derivative of E. coli WP2 under conditions of amino acid starvation and relation to stationary-phage (adaptive) mutation. J Bacteriol. 1998;180:2906–2910. doi: 10.1128/jb.180.11.2906-2910.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang MS, Wang T-CV, Patrick MH. DNA turnover in buffer-held E. coli and its effect on repair of UV damage. Photochem Photobiol. 1979;79:511–520. doi: 10.1111/j.1751-1097.1979.tb07083.x. [DOI] [PubMed] [Google Scholar]
- Rosche WA, Foster PL. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc Natl Acad Sci USA. 1999;96:6862–6867. doi: 10.1073/pnas.96.12.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A, Stahl FW. Double-strand end repair via the RecBC pathway in E. coli primes DNA replication. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Cadwell GW, Barnard KG, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L. Mechanism of induction of adaptive mutations in E. coli. Mol Microbiol. 1990;4:597–601. doi: 10.1111/j.1365-2958.1990.tb00628.x. [DOI] [PubMed] [Google Scholar]
- Bridges BA, Foster PL, Timms AR. Effect of endogenous carotenoids on adaptive mutation in E. coli FC40. Mut Res. 2000;9053:1–11. doi: 10.1016/s0027-5107(00)00144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu Rev Genet. 1999;33:57–88. doi: 10.1146/annurev.genet.33.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Radman M, Smith JM, Toupance B, Gouyon PH, Golelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–2. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- Bridge BA, Timms AR. Mutation in E. coli under starvation conditions: a new pathway leading to small deletions in strains defective in mismatch correction. EMBO J. 1997;16:3349–3356. doi: 10.1093/emboj/16.11.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Maitc I, Radman M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci USA. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski D, Gerrish PJ, kenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The information about HITACHI 835 high speed amino acids analyzer.
The leucine concentration determination conditions.