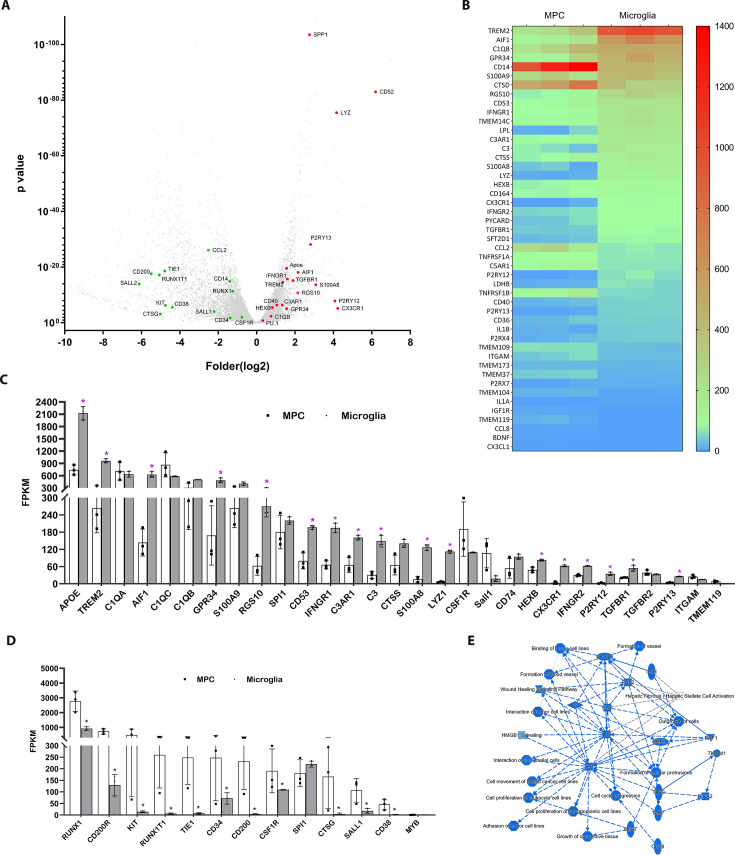
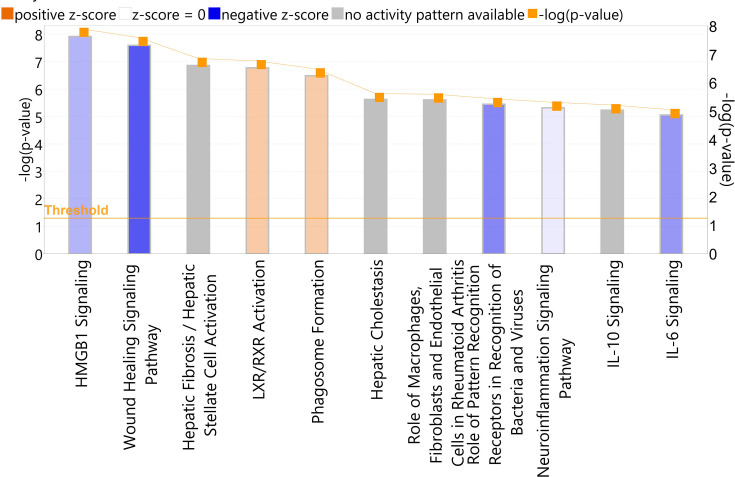
Figure 2. Profiling of genes differentially expression between differentiated microglial cells vs. myeloid progenitor cells (MPCs) using bulk RNAseq analysis.
(A) Volcano plot showing representative genes that were either upregulated (red) or downregulated (green) in differentiated microglia vs. MPCs. (B) Heat map showing increased expression of microglia-enriched genes in differentiated microglia (Supplementary file 1). (C) Histogram comparing the expression levels of microglia-enriched genes in terms of Fragments Per Kilobase of transcript per Million mapped reads (FPKM). *p < 0.05. (D) Histogram comparing expression levels of myeloid cell lineage genes in human-induced pluripotent stem cell (iPSC)-derived MPC and microglia cells using FPKM. *p < 0.05. (E) Graphic signaling pathway analysis with Ingenuity Pathway Analysis (IPA) highlighting IL6 and IL1B as signaling hubs in differential gene expression patterns (Supplementary file 1) in differentiated microglia vs. MPCs.