Figure 4.
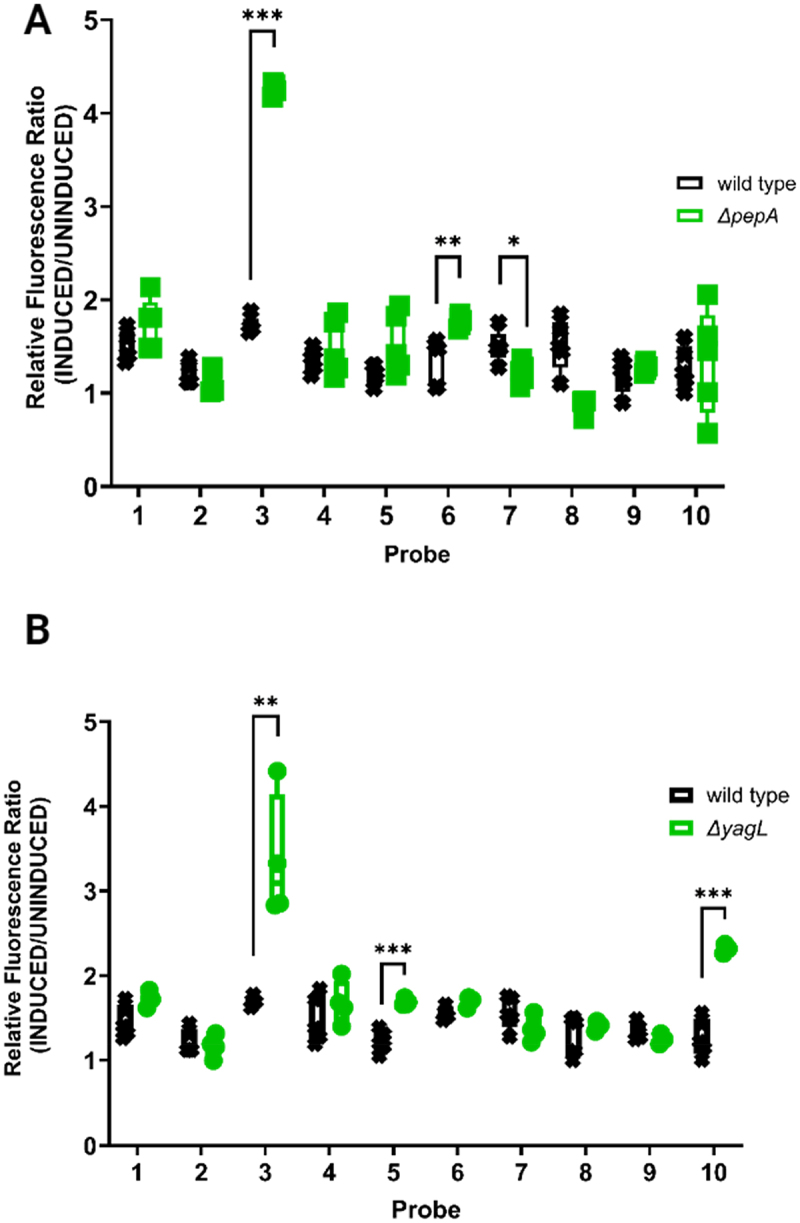
Protein-mediated changes on the accessibility profile of the Tetrahymena ribozyme suggest RNA chaperone role for PepA and YagL. (A) Fluorescence of iRS [3-]targeting of the Tetrahymena ribozyme when expressed in E. coli wild type BW25113 and ΔpepA. For each probe, fluorescence ratios were calculated by dividing paired induced (asRNA probe + ribozyme) by target-uninduced (asRNA probe only) median fluorescence values. Individual data points shown in the graph (‘□’. ΔpepA and ‘x’ -wild type) represent the obtained values for independent biological quadruplets. (B) Ribozyme accessibility profile captured by the iRS [3] assay when expressed in E. coli wild type and ΔyagL. for each probe, fluorescence ratios were calculated by dividing induced (asRNA probe + ribozyme) by uninduced (asRNA probe only) median fluorescence values for independent biological quadruplets (depicted as ‘○’- ΔyagL and ‘x’ -wild type in the graph). Asterisks denote significant difference in the fluorescence ratio for observed in the mutant strain (ΔpepA or ΔyagL) relative to that of the wild-type strain (unpaired t-test; *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001).
