Fig. 4.
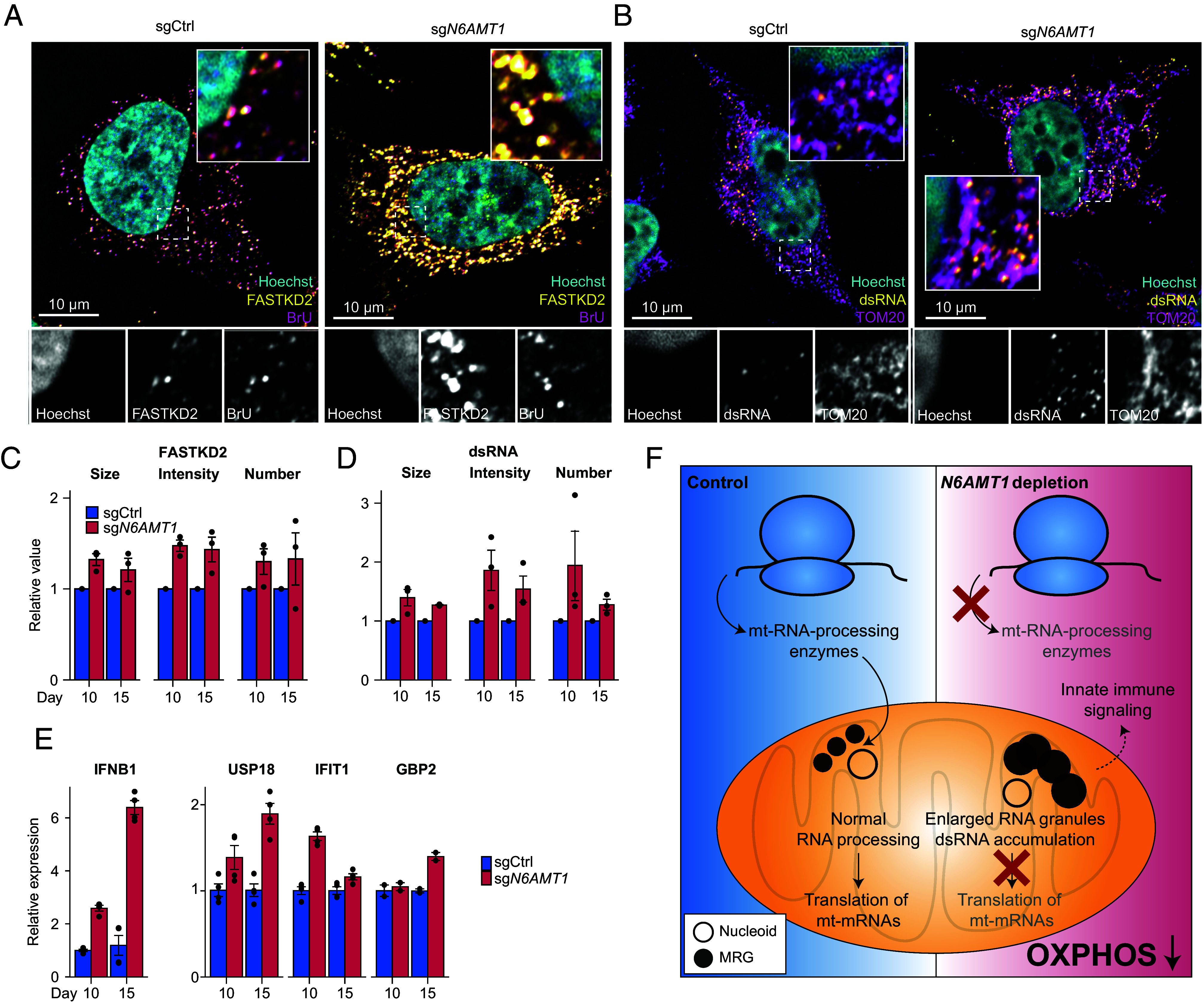
Mitochondrial preRNA and dsRNA accumulate and trigger an interferon response in N6AMT1-depleted cells. (A) Representative images from immunofluorescence analysis of FASTKD2- and bromouridine (BrU)-immunolabeled MRGs in control (sgCtrl) or N6AMT1-depleted (sgN6AMT1) HeLa cells 15 d post transduction. (B) Representative images from immunofluorescence analysis of mitochondria (labeled with antibodies against TOM20) and mitochondrial dsRNA (labeled with J2 dsRNA antibody) in sgCtrl or sgN6AMT1 HeLa cells 15 d post transduction. (C and D) Quantification of FASTKD2 foci (C) and dsRNA foci (D), 10 or 15 d post transduction of HeLa cells with sgCtrl or sgN6AMT1. n = 3 independent experiments, with signal from at least 25 cells per condition. “Number” refers to average number of foci per cell area, “Size” refers to the average foci area, and “Intensity” refers to the average foci intensity. (E) qPCR determination of transcripts of interferon beta (IFNB1) and interferon-inducible USP18, IFIT1, and GBP2 in N6AMT1-depleted HeLa cell lines as compared to controls 10 or 15 d post transduction. n = 4 independent transductions. (F) Proposed role of N6AMT1 in mitochondrial biogenesis. In N6AMT1-depleted cells, translation of mt-RNA-processing enzymes is reduced, leading to accumulation of unprocessed RNA in MRGs. This prevents translation of essential genes involved in mitochondrial gene expression and OXPHOS. In addition, impaired RNA processing leads to the accumulation of double-stranded mt-RNA that activates immune signaling pathways.
