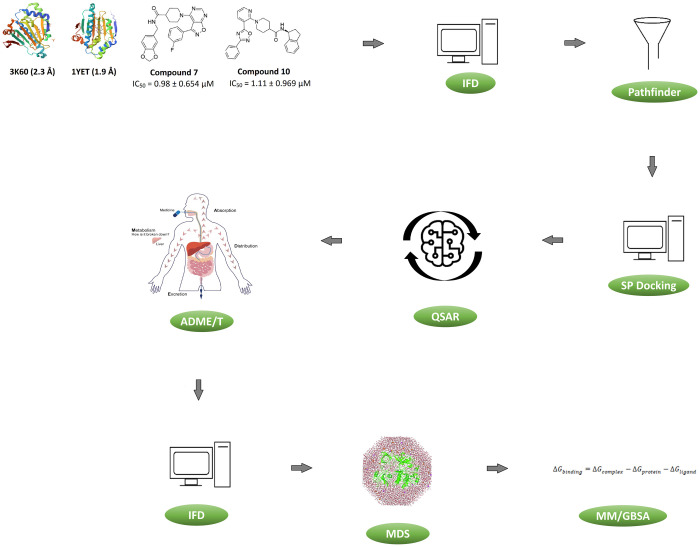
Fig 1. In silico flow diagram showing methodology used on how the reference compounds 10 and 7 were subjected to induced fit docking (IFD) using human Hsp90 (PDB code: 1YET) and PfHsp90 (PDB code: 3K60).
Followed by the selection of compound 10 which was subsequently subjected to pathfinder reaction-based enumeration to yield 10, 000 unique design ideas. These design ideas and compounds from the ChEMBL database were further used to train 13 Auto-QSAR models through reiterative training and testing and scoring of the compounds using standard precision (SP) Glide docking. The adsorption, distribution, metabolism, excretion, and toxicity (ADME/T) were used to filter the compounds, followed by rescoring by IFD and molecular dynamics simulations (MDS) to understand the stability of the protein-ligand complex and conformational changes induced upon ligand binding. The relative binding free energies were estimated by molecular mechanics with generalized born surface area (MM-GB/SA).