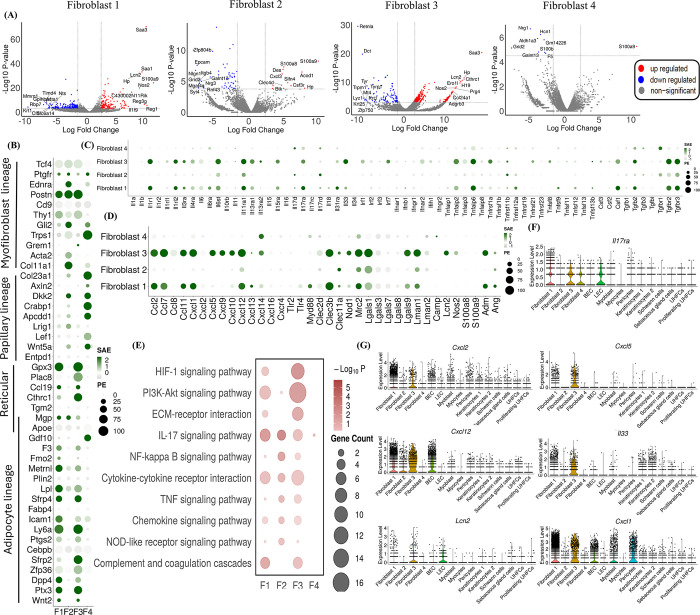
Fig 4. scRNA seq revealed the transcriptomic signatures in fibroblast and other non-immune cells during C. auris skin infection.
A) The Volcano plots indicate significant differentially expressed genes (DEGs) in the infected and uninfected fibroblast subsets, highlighting the top 10 upregulated and downregulated genes. The Dot plot represents the expression of selected B) fibroblast lineage markers, C) cytokines and their receptors, and D) chemokines and their receptors, fungal recognizing receptors, and antimicrobial peptides (AMPs) in the fibroblast subsets (Y-axis). The dot size represents the percentage of cells with expressions, and the color indicates the scaled average expression calculated from the 3 uninfected and 3 infected samples. E) The bubble plot represents the KEGG pathway enrichment of the upregulated genes of fibroblast subsets (X-axis).; Violin plots depict the scaled count of F) IL17ra and G) Cxcl1, Cxcl2, Cxcl5, Cxcl12, Lcn2, and IL33 in fibroblast subsets and other non-immune cells in both uninfected and infected samples. The scaled count were normalized using SCTransform method; DEGs from pseudo-bulk analysis with threshold Log 2-fold change ± 2 and FDR > 5% were considered as significant DEGs and Log 2-fold change ≥ 2 and FDR > 5% as upregulated genes.