Abstract
Tick-borne encephalitis virus (TBEV) is a significant viral pathogen transmitted by ticks, causing severe neurological complications in humans across Europe and Asia, highlighting the urgent need for an in-depth understanding of molecular functions of viral proteins and their interactions with the host proteome. Multi-omics analysis of how TBEV hijack cellular processes provides information about their replication and pathogenic mechanisms. Here, we focused on the proteome, phosphoproteome, and acetylproteome of Vero cells infected by TBEV, revealing the host perturbations triggered by TBEV infection. Additionally, we performed protein-protein interactome analysis to examine the interactions between TBEV and the host. We have provided technical validation, demonstrating the high quality and correlation of samples across all datasets, and evidence of biological consistency of virus-infected cells at the proteomic, phosphoproteomics and acetylomic levels. This comprehensive multi-omics dataset serves as a valuable resource for studying TBEV pathogenesis and identifying potential drug targets for TBEV therapy.
Subject terms: Pathogens, Infection, Protein-protein interaction networks
Background & Summary
Tick-borne encephalitis virus (TBEV) is an arbovirus belongs to the Flaviviridae family, which also includes medically significant viruses such as dengue fever virus (DENV), Zika virus (ZIKV), yellow fever virus (YFV), West Nile virus (WNV), and Japanese encephalitis virus (JEV). TBEV primarily effects the central nervous system and can cause encephalitis and meningitis in severe cases. The virus is predominantly prevalent in Europe and Asia, with approximately 13,000 TBE cases reported each year1. The number of clinically reported TBEV cases is increasing due to the expanding human activities and the wider distribution of ticks2. Although vaccines against TBEV are available, the continuous mutation of the virus poses challenges to the efficacy of these vaccines3,4. Furthermore, there are currently no effective drugs specifically targeting TBEV, suggesting the urgent need to understand the pathogenesis of TBEV and identify potential antiviral targets.
Similar to other flaviviruses, TBEV possesses a single-stranded positive-sense RNA of approximately 11 kb, encoding one open reading frame (ORF) that is processed into three structural proteins: Capsid (C), pre-membrane (prM), Envelope (E), which play crucial roles in virus entry, assembly and release5,6. Additionally, TBEV encodes seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) that facilitate viral RNA replication, protein processing and host immune evasion7–9. Extensive research on the interaction between specific viral proteins and host factors has significantly enhanced our understanding of TBEV’s pathogenesis9–11. However, a comprehensive analysis of the protein-protein interactions (PPI) between TBEV and host is essential to unravel the intricate mechanisms underlying the pathogenesis of TBEV.
Post-translational modifications (PTMs), including methylation, ubiquitination, phosphorylation, acetylation and succinylation, play significant roles in virus pathogenesis and are considered promising targets for antiviral strategies12–15. The intricate interplay between different PTMs contributes extensively to the precision and complexity of cellular regulatory networks16. Simultaneously analyzing different PTMs can examine the coordinated involvement of various regulatory processes during viral infection, thereby gaining a comprehensive understanding of virus pathology and identify valuable targets for antiviral interventions17,18. Additionally, the investigation of virus-host PPI network and PTMs perturbed by virus infection reveals a vast array of host proteins and intricate networks that are manipulated by viral proteins. However, the exploration of multi-proteomics of flavivirus, especially for tick-borne flaviviruses is limited, hindering the comprehensive understanding on the mechanisms by which viral proteins exploit and interact with cellular components.
Here we provide a large dataset of proteome, phosphoproteome and acetylproteome analysis of TBEV-infected Vero cells over a time course, and the analyzed the interactome of between TBEV and host (Fig. 1). This comprehensive assessment of post-translational modifications and protein-protein interactions offers crucial insights into the intricate interplay between the virus and its host, shedding light on the molecular mechanisms of viral pathogenesis. By deciphering these mechanisms, we can gain a deeper understanding of the host’s response to viral infection, potentially facilitating further understanding of the pathogenesis of TBEV and related flaviviruses, and aid in the identification of potential targets for antiviral intervention.
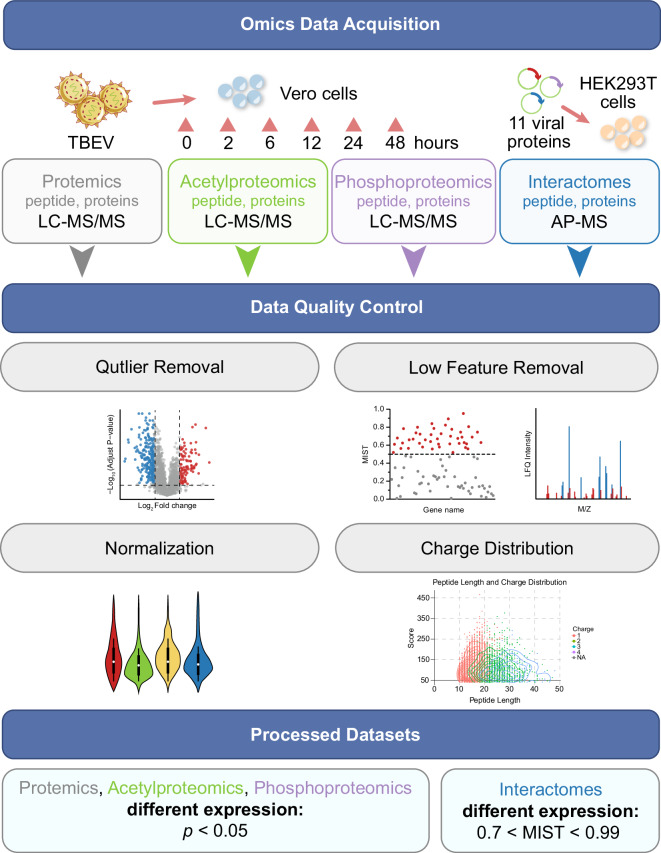
Fig. 1.
This schematic represents the workflow of the multi-proteomics analysis of host perturbations upon TBEV infection and the interactions between TBEV and host. The process starts with the collection of samples from cells infected with TBEV together with cells transfected with viral expression plasmids. Quality control was conducted using qPCR, SDS‒PAGE, and mass spectrometry. The workflow includes detailed analysis of the proteome, acetylproteome, phosphoproteome and interactome to map host responses. These analyses utilize various techniques illustrated by icons and graphs depicting protein modifications and changes over time. This iterative process enables a refined understanding and manipulation of host‒pathogen interactions.
Methods
Proteome, phosphoproteome, and acetylproteome profilings of TBEV-infected cells
Cell line and virus infection
Vero cells (Pricella, CL-0242) were cultured at 37°C with 5% CO2 in DMEM medium (HyClone, Logan, USA). The medium was supplemented with 10% fetal bovine serum (BBI, Shanghai, China) and 1% penicillin-streptomycin (Pen/Strep) (Pricella, Wuhan, China).
The TBEV strain, classified as a member of the Far-Eastern subtype, was isolated in southeast China19. Vero cells were used to propagate the virus, and the titer of the virus was evaluated by tissue culture median infectious dose assay (TCID50). All experiments involving viral infection were conducted at Biosafety Level 3 (BSL-3) laboratory.
For the proteomics analysis, a total of 8 million Vero cells were seeded into T75 flasks. After overnight adherence, the cells were infected with TBEV at a multiplicity of infection (MOI) of 2.0. At various timepoints post-infection (2, 6, 12, 24 and 48 hours), the cells were collected for further analysis. The cells were lysed using SDS lysis buffer (100 mM Tris-HCl pH 8.5; 4% SDS) complemented with protease inhibitor cocktail III, and inhibitors of deacetylation including nicotinamide (NAM) and trichostatin A (TSA). To ensure sufficient material for analysis, cells from seven T75 flasks were pooled together for each sample, which then subjected to analysis for the proteome, phosphoproteome and acetylproteome.
Proteome, phosphoproteome and acetylproteome MS processing
The lysed samples were sonicated on ice for 3 min with the power of 220 W (sonification for 3 seconds followed by a 5-seconds pause). Subsequently, the samples were boiled at 100°C for 30 min to inactivate the virus. After boiling, the samples were centrifuged at 12000 rpm, 14°C for 10 min to pellet the cell debris. The supernatants were carefully transferred to new tubes for concentration assessment using a BCA kit (Thermo, Waltham, USA). Equal amounts of proteins were taken from each sample, and different samples were adjusted to the same volume using lysis buffer. To precipitate proteins, the samples were initially mixed with 1 volume of cold acetone by vertexing. Then, 4 volumes of acetone were added to the samples, which were subsequently precipitated at −20°C for 2 h. After precipitation, the samples were centrifuged at 4,500 g for 5 minutes, and the resulting pellet was washed twice with cold acetone.
For the enrichment of phosphorylated peptides, peptide mixtures were initially incubated with IMAC microspheres suspension in loading buffer (50% acetonitrile/0.5% acetic acid) with vibration. Subsequently, to eliminate non-specifically adsorbed peptides, the IMAC microspheres underwent sequential washing with 50% acetonitrile/0.5% acetic acid and 30% acetonitrile/0.1% trifluoroacetic acid. For elution of the enriched phosphopeptides, an elution buffer containing 10% NH4OH was added, and the enriched phosphopeptides were then eluted with vibration. The supernatant containing phosphopeptides was collected and subsequently lyophilized for liquid chromatography-mass spectrometry (LC-MS) analysis.
For the acetyl peptides enrichment, the peptides were dissolved in IP buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40, pH 8.0) and the supernatant was transferred to pre-washed acetylation resin (Hangzhou PTM Bio-Tech Co., Ltd.), gently shaken at 4 °C overnight. After the incubation, the resin was washed four times with IP buffer and then washed twice with deionized water. Finally, 0.1% trifluoroacetic acid elution buffer was used to elute the bound peptides from the resin, and the elution was performed three times. The eluate was collected and vacuum freeze-dried. After drying, desalting was performed according to the C18 ZipTips instructions, followed by vacuum freeze-drying for LC-MS analysis.
The peptide was dissolved in the liquid chromatography mobile phase A (0.1% formic acid and 2% acetonitrile) and then subjected to separation using the NanoElute ultra-high performance liquid chromatography system. The gradient for liquid phase was set as follows: from 0 to 70 minutes, a gradient of 6% to 24% mobile phase B (0.1% formic acid and 100% acetonitrile) was applied; from 70 to 84 minutes, the gradient of mobile phase B increased from 24% to 35%; from 84 to 87 minutes, the gradient of mobile phase B further increased from 35% to 80%; and from 87 to 90 minutes, the mobile phase B was maintained at 80%, with the flow rate at 450 nL/min. Subsequently, the separated peptide was injected into a Capillary ion source for ionization and further analyzed using the timsTOF Pro mass spectrometry system. The ion source voltage was set to 1.7 kV, and the peptide were detected and analyzed using high-resolution TOF. The scanning range for the secondary mass spectrometry was set from 100 to 1700, and the data collection mode was set as the parallel cumulative serial fragmentation (PASEF) mode. After acquiring a primary mass spectrometry image, 10 PASEF modes were used to collect secondary spectra of parent ions with charge numbers ranging from 0 to 5. To avoid repetitive scanning of parent ions, a dynamic exclusion time of 30 seconds was set for tandem mass spectrometry scanning.
MS quantitative analysis
Raw MS data for proteomics, phosphorylated proteomics, and acetylated proteomics at specific time points during cellular infection (0, 2, 6, 12, 24, and 48 hpi) were processed using MaxQuant (v1.6.17.0) employing default settings (Score > 40, false discovery rate < 1%, and a threshold > 0.75). The protein databases utilized included the Chlorocebus proteome (Blast_Chlorocebus_sabaeus_60711_PR_20210928), the Mycoplasma proteome (UniProt-reviewed_yes + AND + organism_mycoplasma), and targeted TBEV protein databases. A decoy database was incorporated to calculate the false discovery rate (FDR) due to random matches, and a contaminant database was included to minimize the influence of common contaminants on identification results. Trypsin/P was used as the digestion enzyme, allowing for up to two missed cleavages. The minimum peptide length was set to 7 amino acid residues, with a maximum of 5 modifications per peptide. The mass tolerance for precursor ions was set to 20 ppm for both the first search and the main search, with the fragment ion mass tolerance also set to 20 ppm. Carbamidomethylation of cysteine (C) was specified as a fixed modification, while variable modifications included oxidation of methionine, acetylation of the protein N-terminus, and phosphorylation of serine, threonine, and tyrosine. To map the identified Chlorocebus peptides to their corresponding human sequences, we used the R package Biostrings (v. 2.52), leveraging orthologous protein sequences between Chlorocebus and humans.
Differential protein analysis
For the analysis of differential protein expression across multiple time points (0, 2, 6, 12, 24, 48 hpi), the MSstats R package was employed20. This package facilitated comprehensive data processing and analysis. Raw intensity data were log2-transformed to stabilize variance and normalize across samples, with missing values imputed using the package’s default method. A linear mixed-effects model was fitted for each protein, accounting for fixed effects of time points and random effects of biological and technical replicates. To address multiple comparisons, the Benjamini-Hochberg procedure was used to control the FDR at 0.05, identifying proteins with an adjusted p-value below this threshold as significantly differentially expressed.
Statistical analysis
The R packages ggplot2 (https://CRAN.R-project.org/package=ggplot2), dplyr (https://CRAN.R-project.org/package=dplyr), and tidyr (https://CRAN.R-project.org/package=tidyr) were employed to perform a series of quality control evaluations to ensure the integrity of the data21. Specifically, dplyr and tidyr were utilized for data manipulation tasks, including filtering, selecting, grouping, and reformatting. ggplot2 was then used for subsequent data visualization and statistical analysis.
TBEV-host protein-protein interaction AP-MS methods
Cell line and plasmids
HEK293T cells (Pricella, CL-0005) were cultured in DMEM supplemented with 10%FBS and 1% Pen/Strep at 37°C with 5% CO2. Prior to use, all cells were thoroughly tested and confirmed to be free of mycoplasma contamination.
Plasmids encoding ten TBEV structural and non-structural proteins were synthesized by Sangon Biotech (Shanghai, China). The synthesis process involved the generation of nucleotides, which were then inserted into a Flag-VR1012 vector incorporating a 2 × Strep II affinity and 3 × Flag tags at the N-terminal of the coding sequences, the detailed nuclear acid and amino acid sequences for each viral protein were provided in Table S1.
Transfection of plasmids, sample collection and affinity purification
For transfection of HEK293T cells, a total of 10 million HEK293T cells were seeded into a 15 cm petri dish. After allowing the cells to adhere for 24 hours, 25 μg plasmids encoding viral proteins were transfected into cells using PEI transfection reagents (Yeason, China) in a serum-free medium. After 4–6 hours, the medium was removed, and replaced with DMEM supplemented with 10% FBS and 1% Pen/Strep. Following additional 48 hours incubation, the culture medium was discarded, and the cells were collected using cold PBS. The cell pellets were obtained by centrifuging at 12,000 rpm for 1 minutes. The supernatants were completely discarded, and the cell pellets were immediately lysed for protein purification or frozen at −80°C for further use.
The collected cells were lysed using 5 ml immunoprecipitation (IP) buffer containing 50 mM Tris (pH 7.5), 1 mM EGTA, 1 mM EDTA, 1% Triton X-100 and150 mM NaCl supplemented with protease inhibitors (Selleck, Houston, USA). The samples were incubated at 4°C for 30 minutes, and then centrifuged at 12,000 rpm and 4 °C for 15 minutes to remove cell debris. The resulting supernatants were collected for further use.
To purify the viral proteins and their interacting host proteins, Strep-Tactin® XT Purification column (IBA Lifesciences, Germany) was used. The column was first equilibrated with 2 × 1 ml Buffer W (100 mM Tris PH8.0, 150 mM NaCl, 1 mM EDTA). The cell lysates extract was applied to the column and washed with 5 × 1 ml buffer W. The protein of interest, which were expected to be bound to the column, were eluted using 6 × 0.5 ml buffer BXT (100 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 50 mM biotin). Subsequently, the eluted proteins were concentrated using Amicon Ultra-4 (Millipore, USA). All purification steps were carried out at 4 °C. The concentration of primary and the purified proteins were determined using pierce BCA protein assay kit (Invitrogen).
Mass spectrometry analysis
The mass spectrometry analysis of virus-interacting proteins was performed at the National Facility for Protein Science in Shanghai (Shanghai, China). The enriched proteins were first subjected to reduction using 10 mM dithiothreitol (DTT). Subsequently, alkylation was performed using 55 mM iodoacetamide (IAM) to prevent reformation of disulfide bonds. The proteins were then digested into peptide fragments and analyzed using the liquid chromatography System (EASY-nLC 1200, Thermo Scientific) coupled with the Orbitrap Exploris 240 Mass Spectrometer (Thermo Scientific).
During the liquid chromatography-mass spectrometry (LC-MS) analysis, the mobile phase consisted of two solvents: solvent A (0.1% formic acid (FA) in water) and solvent B (0.1% FA in 80% acetonitrile). The peptide samples were loaded onto the trap column at a flow rate of 5 µl/min, and then separated on the analytical column. The mass spectrometer was operated in positive ion mode, utilizing a standard data-dependent acquisition method. The Advanced Peak Detection function was enabled to identify and select the top 20 ions for fragmentation. Precursor ion fragmentation was achieved High Collision Dissociation (HCD) with a normalized collision energy of 30%. For mass analysis, the Orbitrap mass analyzer was set to a resolution of 60,000 for MS1 and 15,000 for MS2. Full scan MS1 spectra were collected in the mass range of 350-1,400 m/z (mass-to-charge ratio), with an isolation window of 1.2 m/z and a fixed first mass of 100 m/z for MS2. Other instrument settings included a spray voltage of 2.2 kV, an Automatic Gain Control (AGC) of 300% for MS1 and the standard setting for MS2, respectively. The maximum ion injection time was set to 50 ms for MS1 and 110 ms for MS2, allowing sufficient time for accurate mass measurements and fragmentation analysis.
MS quantitative analysis
Raw AP-MS data were performed by MaxQuant (v1.6.17.0) with default with default values. For search and quantification of protein, all MS data were processed using the standard Andromeda parameters, incorporating methionine oxidation as a variable modification and carbamidomethylation of cysteine as a fixed modification. Protein identification and peptide-spectrum match (PSM) validation were stringently controlled at a 1% false discovery rate (FDR). All AP-MS data were searched against uniprot-filtered human sequences (20,407 proteins, downloaded on May 25, 2021) and uniprot-reviewed mycoplasma sequences (3,040 proteins, downloaded on May 25, 2021).
Analysis of TBEV-host protein-protein interaction
In the evaluation of affinity purification and mass spectrometry (AP-MS) data, label-free quantification (LFQ) intensity is utilized as the primary measure for quantifying proteins, facilitating comparisons between experimental and control groups. The mass spectrometry interaction statistics (MiST) algorithm was used to rigorously analyze each MS dataset, based on protein abundance, specificity, and technical and biological replicates22. The MiST algorithm assigns a score to each identified protein, predicting its interacting proteins (https://github.com/everschueren/MiST)23. Proteins with a MiST score of ≥0.7 in TBEV bait‒prey experiments were identified as proteins that specifically interact with TBEV24,25.
Data Records
All mass spectrometry data were uploaded to the iProX partner repository with the dataset identifier PXD04843926. This dataset includes raw data from proteome, acetylproteome, phosphoproteome, and interactome. There are a total of 4 raw data folders: proteomics, acetylproteome, and phosphoproteome each contain 20 files, while interactome contains 44 files. Each dataset also includes a MaxQuant search output file, proteinGroups.txt, which contains all the protein groups found by searching the associated raw files.
Technical Validation
Successful infection of virus and expression of viral proteins
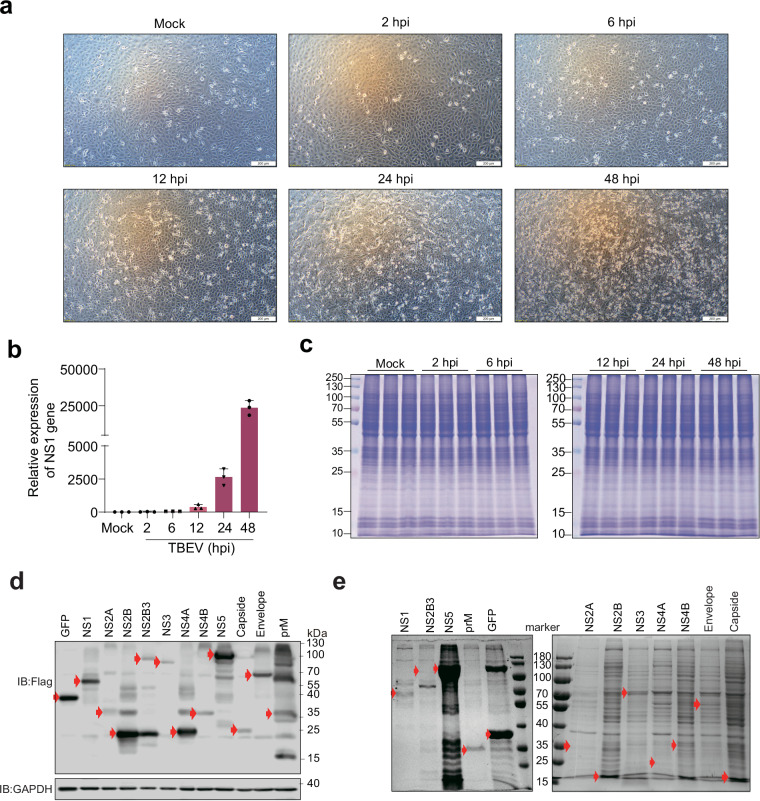
We conducted proteomic, phosphoproteomic, and acetylmic analyses of TBEV-infected cells at various timepoints, with mock-infected cells serving as the control group (Fig. 2). At 12 hours post infection (hpi), subtle cytopathic effects (CPE) were observed, which became more pronounced at 24 hpi and severe at 48 hpi (Fig. 3a). Consistent with the observed CPE in TBEV-infected cells, the mRNA level of the NS1 gene of TBEV was also increased with the duration of virus infection (Fig. 3b). Importantly, as we prepared triplicate samples for each timepoint, equal amounts of proteins in each sample were used for proteome, phosphoproteome and acetylproteome analysis (Fig. 3c). Furthermore, the expression of viral proteins and the efficiency of purification were verified using immunoblot assay and Coomassie brilliant blue stain, respectively (Fig. 3d,e).
Fig. 2.
This diagram presents a workflow for processing multi-proteomics data, including proteomics, acetylproteomics, phosphoproteomics, and interactome data. The process begins with data acquisition using LC‒MS/MS and AP-MS techniques, followed by rigorous quality control steps such as outlier removal, low feature removal, and normalization. The quality control steps are illustrated with various plots depicting the statistical methods used to ensure data integrity. Following data processing, statistically significant datasets were identified with specific thresholds (p < 0.05 for proteomics and 0.5 < MIST < 0.99 for interactome) for further analysis.
Fig. 3.
Successful infection of Vero cells with TBEV. (a) Changes in cell morphology were observed using an optical microscope to observe the morphology of cells infected with TBEV at different time points. (b) Measurement of NS1 gene expression levels at different time points via quantitative PCR (qPCR). (c) Protein gel electrophoresis (SDS‒PAGE) was utilized to observe changes in the expression of total proteins upon TBEV infection. (d) Viral baits expressed in HEK293T cells and analyzed by immunoblot assay. (e) Following affinity purification, enrichment was verified by Coomassie brilliant blue stain analysis. The expected bands for each prey were highlighted in red arrow.
Detection of peptides and proteins
In the proteomic analysis, we were able to detect tryptic peptides in each sample at a 1% false discovery rate (FDR) cut-off. The total number of spectra generated by mass spectrometry in proteomics analysis was 3,212,294, out of which 815,733 spectra matched with theoretical second-order spectra. We identified a total of 62,009 peptides and 6,077 proteins (Table S2). In the phosphoproteomic analysis, the total number of spectra obtained was 2,669,511. We identified 17,300 phosphorylated sites on serine, threonine, and tyrosine, as well as 4,483 phosphorylated proteins. Furthermore, in the acetylomic analysis, we identified 2,844 acetylated proteins and 7,063 acetylated lysine sites out of a total of 1,129,915 spectra (Tables S3, S4).
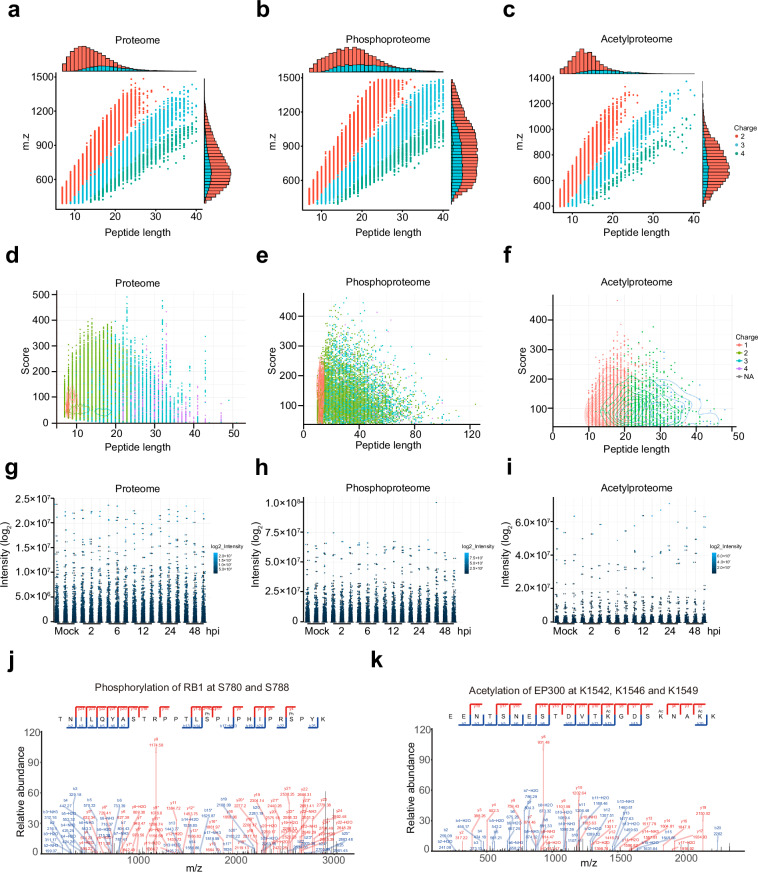
The length of most peptides in proteome is concentrated between 10 to 30 amino acids, exhibiting a diverse distribution across different charge states (Fig. 4a,d). The highest number of peptides corresponds to charge 2, and the two-dimensional density distribution map depicting their length and score reveals a high-density region, indicating that these peptides possess good quality. Although the number of peptides with charge 3 and 4 is relatively small, their scores are relatively high. In contrast, the peptides involved in phosphorylation modifications exhibit a wider range of lengths, with some peptides exceeding 100 amino acids (Fig. 4b,e). Phosphorylation modifications typically occur at serine, threonine, or tyrosine residues, which may be distributed at different positions, resulting in longer peptide lengths. Regarding acetylation, the length of peptides is mainly concentrated between 10 to 40 amino acids (Fig. 4c,f). Acetylation modifications predominantly occur at the N-terminus or lysine residues of proteins, leading to relatively concentrated peptide lengths. The distribution of peptide intensities in the proteome appears relatively consistent, with a few noticeable outliers indicating significantly high expression levels of certain proteins in specific samples (Fig. 4g-f). Overall, the graph provides a comprehensive view of the length, score, and charge state of peptide, indicating good quality of the data.
Fig. 4.
Detection of peptides in individual runs. (a–c) The relationship between peptide length and its corresponding mass-to-charge ratio (m/z). Most peptides of the proteome, acetylproteome, and phosphoproteome identified are distributed between 7 and 20 amino acids in length, aligning with the typical patterns expected from enzymatic digestion and mass spectrometry fragmentation methods. (d–f) Differences in peptide length, score, and charge state among the proteome, acetylproteome, and phosphoproteome data. (g–i) The distribution of peptide intensity values for each sample in the proteome, acetylated proteome, and phosphorylated proteome is presented. (j,k) The results of protein mass spectrometry spectra for the indicated phosphorylated (j) and acetylated proteins (k).
Notably, we successfully detected conventional phosphorylation sites, such as phosphorylation of RB1 at S780 and S788, STAT1 at S727, and CDK2 at Y15 and T1427–29 (Fig. 4j and S1a), together with the acetylation of EP300 at K1542, K1546 and K1549, as well as acetylation of ACLY at K540 and K554, and XRCC6 at K55630,31,32 (Fig. 4k and S1b). Additionally, we observed decreased phosphorylation levels of SAMHD1 at S18 and S278 following TBEV infection, a phenomenon also observed during HIV and human cytomegalovirus (HCMV) infection33,34 (Fig. S1a). These collective findings underscore the credibility of our analysis.
Reproducibility of the multi-proteomics analysis
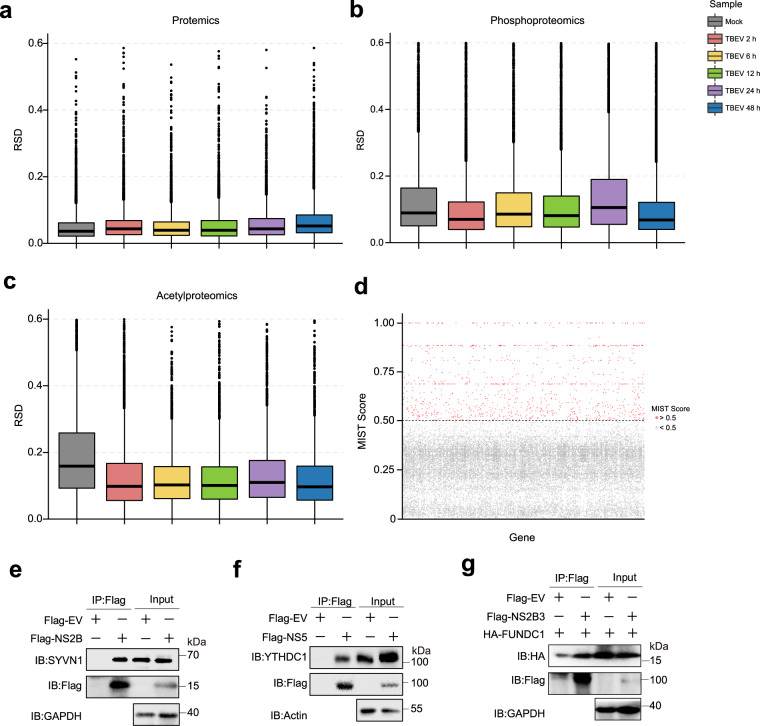
The statistical consistency of proteomics data from the three replicates for each sample was evaluated using the Relative Standard Deviation (RSD). The analysis revealed that all proteomics samples possessed RSD values below 0.2, exhibiting a high level of consistency among replicates (Fig. 5a–c). Among the proteomics results, the replicates showed the best consistency, as they had the lowest RSD values. However, it is worth noting that in the acetylproteome analysis, the mock group displayed the highest RSD value. This observation could be attributed to the low levels of protein acetylation, which were below the detection threshold in some cells following the treatment. Consistently, Spearman’s correlation analysis revealed high consistency between replicates, with correlation coefficients ranging between 0.96 and 1.0 for the proteomics analysis, and 0.86–1.0 for the phosphoproteomics and acetylproteomics analyses (Fig. S2).
Fig. 5.
Repeatability of proteomics results. (a–c) The statistical consistency of proteomics data was evaluated using the Relative Standard Deviation (RSD) analysis. The RSD values were calculated for each sample’s three replicates. (d) The results of interactome analysis based on the MIST score, with each point representing a gene. Red dots indicate high-confidence protein interactions (MIST score greater than 0.5), while gray dots indicate lower-confidence interactions (MIST score less than 0.5). (e–g) Validation of virus-host interactions using co-immunoprecipitation. Flag-tagged empty vector (EV) or viral proteins were overexpressed in HEK293T cells, and endogenous SYVN1 co-precipitated with NS2B (e), YHTDC1 co-precipitated with NS5 (f), along with HA tagged FUNDC1 co-precipitated with NS2B3 (g), were detected using flag beads and immunoblot assay.
In the interactomics analysis, we obtained a total of 1,080 hits with a mist value greater than 0.5 out of the 30,677 identified proteins (Fig. 5d and Table S5). Among these hits, we identified 40.4% (436/1080) of proteins with a mist value between 0.5 and 0.6. Additionally, 24.9% (269/1080) of the interacted proteins showed a mist value between 0.6 and 0.7. The remaining 34.7% (375/1080) of the interacted host proteins possessed a mist value ranging from 0.7 to 1.0. Additionally, the interactions of NS2B-SYVN1, NS5-YTHDC1, and NS2B3-FUNDC1 were further validated by co-immunoprecipitation analysis (Fig. 5e–g), thereby affirming the credibility of our analysis.
Usage Notes
Emerging and re-emerging viral diseases transmitted by arthropods continue to pose a significant threat to both humans and animals, with the potential for severe social and economic consequences. Understanding how viruses perturb host proteins is crucial for researching the pathogenic mechanisms of viral infections. The dataset presented in this study represents the most comprehensive proteomics analysis of TBEV infection and the TBEV-host Protein-Protein Interaction (PPI) network. This dataset has the potential to enhance our understanding of TBEV pathogenesis and related flaviviruses, as well as contribute to the identification of potential targets for antiviral interventions.
Supplementary information
Acknowledgements
This work was supported by grant from the National Natural Science Foundation of China (Grant No. 82372250, 82302516, and 82341105), Natural Science Foundation of Jilin Province (Grant No. YDZJ202301ZYTS431), Scientific Research Project of Jilin Provincial Department of Education (Grant No. JJKH20231206KJ), the Medical Innovation Team Project of Jilin University (2022JBGS02), Jilin Provincial Scientific and Technological Development Program (Grant No. 20240207010CX, State Key Laboratory for Diagnosis and Treatment of Severe Zoonotic Infectious Diseases (Grant No. 2024ZZ00002 & RCGHCRB202405), and Norman Bethune Plan Project of Jilin University (Grant No. 2024B29).
Author contributions
L.S., X.G. and W.W. generated and analyzed the data, wrote the code and led the writing of the manuscript. Y.X. drafted the diagrams. Q.L., Y.Z. and L.S. conceived the idea, supervised the project and contributed extensively to all ideas and critically revised the manuscript.
Code availability
No custom code was made for the compilation and validation procedures in this dataset.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liyan Sui, Email: suiliyan@jlu.edu.cn.
Yicheng Zhao, Email: yichengzhao@live.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04036-y.
References
- 1.Dobler, G., Gniel, D., Petermann, R. & Pfeffer, M. Epidemiology and distribution of tick-borne encephalitis. Wien Med Wochenschr162, 230–238 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Worku, D. A. Tick-Borne Encephalitis (TBE): From Tick to Pathology. J Clin Med12 (2023). [DOI] [PMC free article] [PubMed]
- 3.Dobler, G. et al. Tick-borne encephalitis virus vaccination breakthrough infections in Germany: a retrospective analysis from 2001 to 2018. Clin. Microbiol. Infect.26, 1090.e1097–1090.e1013 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Dai, X., Shang, G., Lu, S., Yang, J. & Xu, J. A new subtype of eastern tick-borne encephalitis virus discovered in Qinghai-Tibet Plateau, China. Emerg Microbes Infect7, 74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraldo, M. I. et al. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature585, 414–419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gestuveo, R. J. et al. Analysis of Zika virus capsid-Aedes aegypti mosquito interactome reveals pro-viral host factors critical for establishing infection. Nat Commun12, 2766 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang, Q. et al. Tick-borne encephalitis virus NS4A ubiquitination antagonizes type I interferon-stimulated STAT1/2 signalling pathway. Emerg Microbes Infect9, 714–726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sui, L. et al. Flavivirus prM interacts with MDA5 and MAVS to inhibit RLR antiviral signaling. Cell Biosci13, 9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gracias, S. et al. Tick-borne flavivirus NS5 antagonizes interferon signaling by inhibiting the catalytic activity of TYK2. EMBO reports24, e57424 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang, J. et al. Sterile 20-like kinase 3 promotes tick-borne encephalitis virus assembly by interacting with NS2A and prM and enhancing the NS2A-NS4A association. Journal of medical virology95, e28610 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Serman, T. et al. Acetylation of the NS3 helicase by KAT5γ is essential for flavivirus replication. Cell Host Microbe31, 1317–1330.e1310 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, Q. et al. The global succinylation of SARS-CoV-2-infected host cells reveals drug targets. Proc Natl Acad Sci USA119, e2123065119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouhaddou, M. et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell182, 685–712.e619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaturro, P. et al. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature561, 253–257 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Xu, G. et al. Multiomics approach reveals the ubiquitination-specific processes hijacked by SARS-CoV-2. Signal Transduct Target Ther7, 312 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stukalov, A. et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature, (2021). [DOI] [PubMed]
- 17.Pinto, S. M. et al. Multi-OMICs landscape of SARS-CoV-2-induced host responses in human lung epithelial cells. iScience26, 105895 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan, J. F., Yuan, S., Chu, H., Sridhar, S. & Yuen, K. Y. COVID-19 drug discovery and treatment options. Nat Rev Microbiol, (2024). [DOI] [PubMed]
- 19.Li, X. et al. Molecular detection and phylogenetic analysis of tick-borne encephalitis virus in ticks in northeastern China. Journal of medical virology94, 507–513 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Choi, M. et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics30, 2524–2526 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Shen, X. et al. TidyMass an object-oriented reproducible analysis framework for LC-MS data. Nat Commun13, 4365 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jager, S. et al. Global landscape of HIV-human protein complexes. Nature481, 365–370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verschueren, E. et al. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr Protoc Bioinformatics49, 8.19.11–18.19.16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baggen, J., Vanstreels, E., Jansen, S. & Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat Microbiol6, 1219–1232 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Liu, Q. & Guo, X. iProX https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD048439 (2024).
- 27.Ramsauer, K. et al. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc Natl Acad Sci USA99, 12859–12864 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes, B. T., Sidorova, J., Swanger, J., Monnat, R. J. Jr. & Clurman, B. E. Essential role for Cdk2 inhibitory phosphorylation during replication stress revealed by a human Cdk2 knockin mutation. Proc Natl Acad Sci USA110, 8954–8959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konagaya, Y., Rosenthal, D., Ratnayeke, N., Fan, Y. & Meyer, T. An intermediate Rb–E2F activity state safeguards proliferation commitment. Nature631, 424–431 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emmons, M. F. et al. HDAC8-mediated inhibition of EP300 drives a transcriptional state that increases melanoma brain metastasis. Nat Commun14, 7759 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, R. et al. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell51, 506–518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen, H. Y. et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell13, 627–638 (2004). [DOI] [PubMed]
- 33.Badia, R. et al. SAMHD1 is active in cycling cells permissive to HIV-1 infection. Antiviral Res142, 123–135 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Kim, E. T. et al. SAMHD1 Modulates Early Steps during Human Cytomegalovirus Infection by Limiting NF-kappaB Activation. Cell Rep28, 434–448 e436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No custom code was made for the compilation and validation procedures in this dataset.