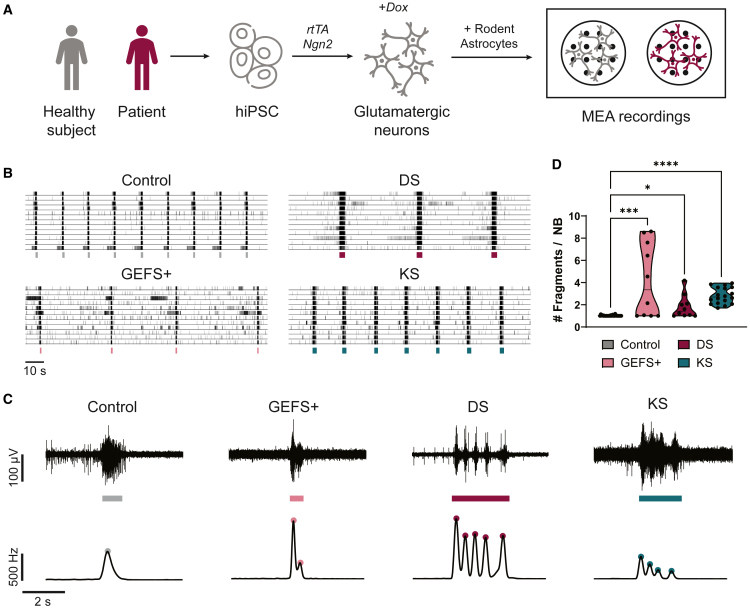
Figure 1.
Fragmented network bursts in patient-derived neuronal networks on MEA
(A) Schematic overview of the protocol used by Van Hugte et al., (2023); Frega et al., 2019 to differentiate hiPSCs into neuronal networks on multi-electrode arrays (MEAs). hiPSCs were obtained by reprogramming somatic cells of healthy subjects and patients. Excitatory neurons were generated through doxycycline (Dox)-inducible overexpression of Neurogenin2 (Ngn2). At day in vitro (DIV) 2, E18 rat astrocytes were added in a 1:1 ratio. Activity was recorded at DIV 35 with MEA.
(B) Representative raster plots of spontaneous activity of healthy control neuronal networks (Control) and networks derived from patients with GEFS+, Dravet Syndrome (DS), and Kleefstra Syndrome (KS) recorded by Van Hugte et al., (2023); Frega et al., 2019. Detected network bursts (NBs) are indicated by the colored bars below.
(C) Top: representative example voltage traces recorded at one electrode during an NB detection (bar below). Bottom: representative network firing rate traces during the same NB and detected fragments (colored dots).
(D) Quantification of the average number of fragments per NB for Control (n = 26), GEFS+ (n = 10), DS (n = 11), and KS (n = 18) networks. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Kruskal-Wallis test with Dunn’s multiple comparisons test was performed between groups.