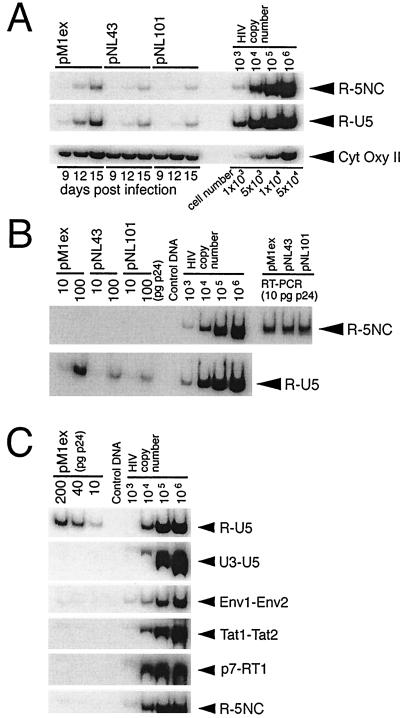
FIG. 6.
PCR analysis of HIV-1 DNA. (A) PCR analysis of HIV-1 DNA in cytosolic fractions of infected cells. Cytosolic DNA was isolated from infected Jurkat cells at days 9, 12, and 15 postinfection, as described in Materials and Methods, and aliquots corresponding to 104 cells were subjected to quantitative PCR using the R-U5 and R-5NC primer pairs. To monitor the efficiency of cytosolic DNA isolation, the mitochondrial CytOxy II gene was amplified. (B) PCR analysis of virion-associated HIV-1 DNA. Virion-associated nucleic acids were extracted, and aliquots corresponding to 10 and 100 pg of p24 were subjected to quantitative PCR using the R-U5 and R-5NC primer pairs. To analyze the amount of viral genomic RNA in the extracted nucleic acids, samples corresponding to 100 ng of p24 were reverse transcribed with Moloney murine leukemia virus RT, and an aliquot of synthesized viral cDNA, corresponding to 10 pg of p24, was subjected to PCR, using the R-5NC primer pair (shown as RT-PCR in the figure). (C) Aliquots of pM1ex virion-associated nucleic acids, corresponding to 10, 40, and 200 pg of p24, were subjected to quantitative PCR, using the indicated primer pairs. Primer pair R-U5 was designed to detect early RT products synthesized either before or immediately after the first template switch. With primer pairs U3-U5, Env1-Env2, Tat1-Tat2, and p7-RT1, a specific PCR product was expected only if negative-strand DNA of increased length, relative to wild-type, had been synthesized after the first template switch. Primer pair R-5NC was predicted to amplify only late RT products synthesized after the second template switch. Serial 10-fold dilutions of pNL43 plasmid were used as positive controls. PCR products were separated on 6% polyacrylamide gels (0.5× TBE) at 4°C, and then the gels were dried and the bands were visualized on X-ray film.