Abstract
Carotenoid biosynthesis is closely associated with abscisic acid (ABA) during the ripening process of non-climacteric fruits, but the regulatory mechanism that links ABA signaling to carotenoid metabolism remains largely unclear. Here, we identified two master regulators of ABA-mediated citrus fruit coloration, CsERF110 and CsERF53, which activate the expression of carotenoid metabolism genes (CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, CsZEP, and CsNCED2) to facilitate carotenoid accumulation. Further investigations showed that CsERF110 not only activates the expression of CsERF53 by binding to its promoter but also interacts with CsERF53 to form the transcriptional regulatory module CsERF110–CsERF53. We also discovered a positive feedback regulatory loop between the ABA signal and carotenoid metabolism regulated by the transcriptional regulatory module CsERF110–CsERF53. Our results reveal that the CsERF110–CsERF53 module responds to ABA signaling, thereby orchestrating citrus fruit coloration. Considering the importance of carotenoid content for citrus and many other carotenoid-rich crops, the revelation of molecular mechanisms that underlie ABA-mediated carotenoid biosynthesis in plants will facilitate the development of transgenic/gene-editing approaches, further contributing to improving the quality of citrus and other carotenoid-rich crops.
Key words: citrus, abscisic acid, ABA, fruit coloration, carotenoid metabolism, ethylene responsive factor, ERF, transcriptional regulatory module
This study reports that abscisic acid activates the transcriptional regulatory module CsERF110–CsERF53 in citrus during ripening process, thus positively regulating carotenoid accumulation.
Introduction
Carotenoids are a type of isoprenoid and one of the most important pigments in nature (Nisar et al., 2015). Since the first isolation of carotenoids from the carrot root in 1831, numerous studies have been performed to investigate their physicochemical properties (Miller et al., 1935). Carotenoids endow plants with multiple colors and function in numerous biological processes such as photosynthesis, photoprotection, antioxidation, and pollination (Nisar et al., 2015; Sandmann, 2021). α-Carotene and β-carotene, well-known carotenoids and vitamin A biosynthetic precursors, are important dietary nutrients, contributing to visual protection, immunoenhancement, disease prevention, and delay of senility in the human body (Saari, 2016; Takahashi et al., 2022). Overall, carotenoids play important roles in physiological and biochemical processes in humans and plants.
In recent decades, the biosynthetic pathways of carotenoids have been well characterized (Lu and Li, 2008; Vranová et al., 2013; Yuan et al., 2015; Zhou et al., 2022). Geranylgeranyl diphosphate (GGPP), the direct precursor of carotenoid biosynthesis, is produced from the GGPP synthase (GGPPS)-catalyzed condensation of isopentenyl diphosphate and dimethylallyl diphosphate. As the key rate-limiting enzyme, GGPPS determines the precursor supply to initiate carotenoid biosynthesis, thus affecting the total yield of carotenoids. Phytoene synthase (PSY) catalyzes the conversion of GGPP into 15-cis-phytoene, the first carotenoid produced in the carotenoid pathway. After two-step desaturation and three-step isomerization, 15-cis-phytoene is converted into all-trans-lycopene, and carotenoid metabolic flow then branches into the α and β directions through lycopene ε-cyclase (LCYE) and lycopene β-cyclase (LCYB), respectively. The downstream α branch of carotenoid metabolism mainly produces the yellow carotenoid lutein. In the β branch, β-carotene hydroxylase transforms LCYB-produced β-carotene into β-cryptoxanthin and zeaxanthin; the latter may be converted to violaxanthin through the catalytic activity of zeaxanthin epoxidase (ZEP). 9-cis-epoxycarotenoid dioxygenase (NCED), a key rate-limiting enzyme of abscisic acid (ABA) biosynthesis and an enzyme of carotenoid decomposition, uses violaxanthin as a precursor to synthesize the phytohormone ABA.
Transcriptional regulation is an essential and widespread regulatory mechanism in plants, participating in almost all processes, including carotenoid metabolism (Stanley and Yuan, 2019; Liang and Li, 2023). Numerous transcription factors (TFs) have been reported to participate in regulating carotenoid metabolism in plants, including members of the MYB, bHLH, MADS, bZIP, NAC, and HD-ZIP families (Zhu et al., 2017, 2023; Gong et al., 2021; Zhang et al., 2021; Sun et al., 2023, 2024; Yue et al., 2023). The ethylene (ETH)-responsive factor (ERF) subfamily is an essential TF family mainly present in plants. TFs in this subfamily serve as important regulators in many biological and physiological processes such as plant morphogenesis, stress responses, phytohormone signal transduction, and metabolite regulation (Hirota et al., 2007; El-Sharkawy et al., 2009; Wang et al., 2010). ERFs act as a hub in the phytohormone crosstalk network, participating in the regulation and signal transduction of ABA, ETH, brassinolide, cytokinin, and gibberellins (Xie et al., 2019). In plant ripening and senescence processes, ERFs regulate the biosynthesis or signal transduction of ABA and ETH, thus modulating plant development and ripening processes (Xie et al., 2022). Coloration induced by the accumulation of carotenoids is an important sign of plant maturity and senescence, and multiple ERF TFs have been reported to participate in the regulation of coloration. For instance, CsERF061, an ETH-induced ERF TF, activates the promoters of nine key carotenoid pathway genes to promote citrus fruit coloration (Zhu et al., 2021). MdAP2-34 directly binds the promoter of MdPSY2-1 and activates its expression, thus promoting carotenoid biosynthesis in apple (Dang et al., 2021). In Lycium, LbERF5.1 positively regulates carotenoid accumulation by activating the expression of LbCCD4.1 (Zhao et al., 2023). These findings highlight the vital regulatory roles of ERFs in modulating carotenoid biosynthesis.
Citrus is one of the most economically important crops worldwide, with an annual production of approximately 158.5 million metric tons in 2021 (Food and Agriculture Organization of the United Nations, https://www.fao.org/faostat/en/#data/). Citrus fruits are widely used in food processing, beverage production, pharmaceutical production, and other industries; they are also major dietary sources of provitamin A, as they contain abundant carotenoids. Citrus fruits have more diverse carotenoids than other fruits and specifically accumulate single carotenoids. In addition, different citrus germplasms exhibit various regulatory mechanisms of carotenoid metabolism (Zhang et al., 2024). These characteristics make citrus an excellent material for studying the transcriptional regulation of carotenoid metabolism. For example, CrMYB68 negatively regulates CrBCH2 and CrNCED5, thereby suppressing the transformation of α- and β-carotene and ABA biosynthesis in the citrus flavedo (Zhu et al., 2017). CsERF061 activates the promoters of nine carotenoid pathway genes, CsPSY1, CsPDS, CsCRTISO, CsLCYb1, CsBCH, CsZEP, CsNCED3, CsCCD1, and CsCCD4, suggesting the multi-regulation functions of CsERF061 in carotenoid biosynthesis (Zhu et al., 2021). The bHLH TF CsTT8 promotes citrus fruit coloration by regulating the methylerythritol 4-phosphate pathway and the carotenoid synthesis pathway (Sun et al., 2023). CsMPK6 interacts with CsMYC2, forming a jasmonate-induced CsMPK6–CsMYC2 negative feedback loop, ultimately regulating β-citraurin accumulation and fruit coloration in citrus (Yue et al., 2023).
Carotenoid metabolism is mainly regulated by various environmental signals (e.g., light, temperature, and phytohormones), especially phytohormone signal stimulation (Fanciullino et al., 2014; Kim et al., 2018; Gong et al., 2021; Yang et al., 2021; Huang et al., 2022; Yue et al., 2023; Sun et al., 2024). Citrus is a typical non-climacteric fruit, and ABA plays an essential role in its ripening process (Rodrigo et al., 2003; Wu et al., 2014; Romero et al., 2019). ABA is an important inducing factor that promotes the formation of coloration traits (Zhang et al., 2021; Sun et al., 2023). However, the role of AP2/ERF TFs in ABA-mediated carotenoid metabolism is still largely unknown. In the present study, we identified a CsERF110–CsERF53 transcriptional regulatory module and found that it positively regulated ABA-mediated fruit coloration in citrus by establishing a positive feedback loop between ABA signaling and carotenoid metabolism. Our findings provide valuable references for the enhancement of carotenoid accumulation and improvement of fruit quality in citrus and other crops.
Results
CsERF110 may participate in ABA-induced carotenoid accumulation as a nuclear-localized transcriptional activator
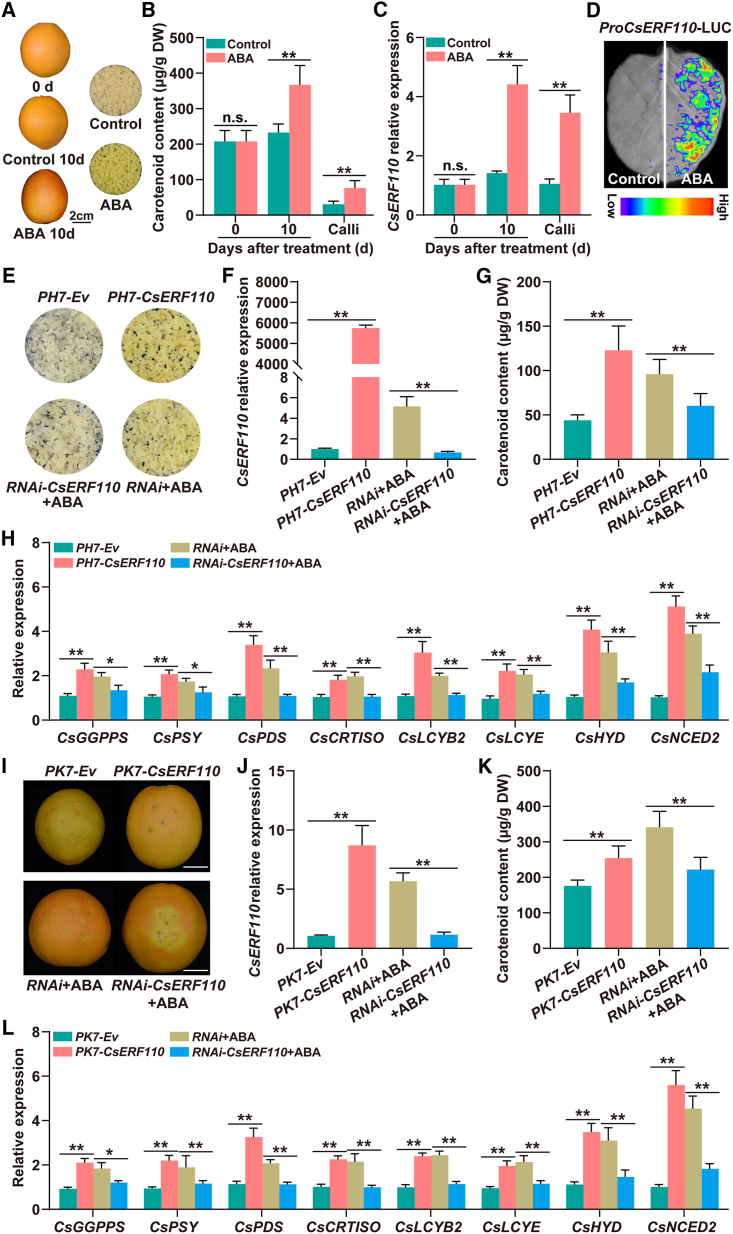
In a previous study, we identified the ABA-induced ERF TF CsERF110 on the basis of RNA sequencing data from ABA-treated calli, and this ERF appeared to exhibit the potential to modulate ABA-mediated carotenoid metabolism (Gao, 2013). In the present study, phylogenetic analysis showed that CsERF110 was clustered with AT1G43160, an ERF protein from Arabidopsis thaliana (Supplemental Figure 1). AT1G43160 has been reported to regulate a variety of biological processes in response to ABA signaling, suggesting that CsERF110 may be involved in regulating ABA-induced carotenoid metabolism in citrus. To test this theory, we performed ABA treatment experiments on citrus fruits and calli. As shown in Figure 1A, on day 10 after treatment, citrus fruits in the control group remained orange, whereas those in the ABA-treated group turned red. Similarly, the calli in the control group remained light white, whereas those in the ABA-treated group turned yellow (Figure 1A). High-performance liquid chromatography (HPLC) results showed that ABA treatment significantly increased the carotenoid content of citrus peel and calli (Figure 1B). Correspondingly, CsERF110 expression was significantly increased in ABA-treated citrus fruits and calli (Figure 1C). We then constructed a ProCsERF110-LUC (luciferase) fusion vector and transiently transformed it into leaves of Nicotiana benthamiana. Higher fluorescence intensity was detected in ABA-treated regions than in water-treated regions (Figure 1D). These results indicated that CsERF110 was induced by ABA signaling and might be involved in regulating ABA-mediated carotenoid accumulation in citrus.
Figure 1.
CsERF110 is essential for ABA-mediated carotenoid biosynthesis in citrus.
(A–C) Phenotypes of citrus fruit and calli (A), (B) carotenoid content, and (C)CsERF110 expression under ABA treatment. Scale bars, 2 cm. (D)In vivo firefly luciferase (LUC) complementation imaging of N. benthamiana leaves treated with water or ABA. A suspension of Agrobacterium GV3101 carrying ProCsERF110-LUC was injected into N. benthamiana leaves. Two days after injection, N. benthamiana leaves were treated with water or 100 μM ABA, and 24 h later, bioluminescence imaging was used to measure the luciferase activity of ProCsERF110.
(E)–(H) Stable transformation of CsERF110 in citrus calli. (E) Phenotypes. PH7-CsERF110 and RNAi-ERF110 indicate CsERF110 overexpression and RNA interference, respectively. The PH7 (PH7-Ev) and RNAi empty vectors are controls. The expression levels of CsERF110(F) and CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, and CsNCED2(H) are shown. (G) Total carotenoid content (μg/g dry weight [DW]).
(I, J, and L) Transient expression of CsERF110 in citrus fruit. (I) Phenotypes. The PK7 (PK7-Ev) and RNAi empty vectors are controls. PK7-CsERF110 and RNAi-CsERF110 indicate CsERF110 overexpression and RNA interference, respectively. Scale bars, 2 cm. The transcript levels of CsERF110(J) and CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, and CsNCED2(L) are shown.
(K) Total carotenoid content (μg/g DW). Data are presented as means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).
To examine the subcellular localization of CsERF110, we constructed the fusion vector GFP-CsERF110 and transiently co-transformed it into N. benthamiana leaves together with a nuclear marker, nuclear transcription factor (NF)-YA-mCherry. Green fluorescence (from GFP-CsERF110) and red fluorescence (from NF-YA-mCherry) co-localized in the nucleus, indicating that CsERF110 was localized in the nucleus (Supplemental Figure 2A). We then examined the transcriptional activity of CsERF110 using the yeast hybrid and dual luciferase systems. In the yeast hybrid system, all transformed yeast strains grew well on SD/-Trp selective medium, but only the BD-CsERF110-transformed strain and the positive control grew well on SD/-Trp/-His/-Ade selective medium (Supplemental Figure 2B). In the dual luciferase system, pBD-CsERF110, pBD (negative control), and pBD-VP16 (positive control) were co-transformed into N. benthamiana leaves (Supplemental Figure 2C). The LUC/REN ratio was significantly higher in the pBD-CsERF110 and pBD-VP16 groups than in the pBD group (Supplemental Figure 2D). These results suggest that ABA-induced CsERF110 functions as a nuclear-localized transcriptional activator.
CsERF110 is involved in ABA-induced carotenoid accumulation as an essential activator in citrus
To identify the function of CsERF110, we performed CsERF110 stable transformation and transient transformation experiments in citrus calli and fruits, respectively (Figure 1E–1L). Transgenic calli with the PH7 and RNAi empty vectors were used as controls for CsERF110-overexpressing calli and CsERF110-interfering calli, respectively. Fruits transiently infected with PK7 and RNAi empty vectors were used as controls for CsERF110-overexpressing fruits and CsERF110-interfering fruits, respectively. The calli in the control group had a pale appearance, whereas the CsERF110-overexpressing calli had a distinct yellowish color (Figure 1E and 1F). Similarly, the peels of CsERF110-overexpressing fruits had a more obvious orange-red color than those of the controls (Figure 1I and 1J). HPLC results showed that the carotenoid content was significantly higher in CsERF110-overexpressing calli and fruits than in their controls (Figure 1G and 1K), thus explaining the differences in coloration of transgenic calli and fruits between the CsERF110 transformation group and the control group (Figure 1E and 1I). To decipher the molecular mechanism underlying the changes in carotenoid content of transgenic calli and fruits, we examined the expression levels of carotenoid metabolism genes using real-time quantitative PCR. As shown in Figure 1H and 1L, CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, and CsNCED2 were significantly upregulated in CsERF110-overexpressing calli and fruits compared with those of the control groups. These results indicate that CsERF110 positively regulates carotenoid accumulation in citrus calli and fruits.
To further explore the role of CsERF110 in ABA-mediated carotenoid synthesis in citrus, we treated CsERF110-RNAi calli and fruits with ABA. As shown in Figure 1E, after ABA treatment, the control calli exhibited a yellowish color, whereas the CsERF110-RNAi calli maintained a light color. Similarly, the peel near the injection pole of ABA-treated CsERF110-RNAi fruits remained yellow, whereas that of the control group turned orange (Figure 1I). In addition, after ABA treatment, the expression of CsERF110 was significantly lower in CsERF110-RNAi calli and fruits than in those of the control (Figure 1F and 1J). HPLC results showed that carotenoid content was significantly higher in ABA-treated control calli and fruits than in the CsERF110-RNAi group (Figure 1G and 1K). Correspondingly, the expression levels of carotenoid metabolism genes were significantly higher in ABA-treated control calli and fruits than in those of CsERF110-RNAi (Figure 1H and 1L). Together, these results demonstrate that CsERF110 acts as an essential activator of ABA-induced carotenoid accumulation by activating the expression of carotenoid biosynthetic genes in citrus.
CsERF110 directly activates the expression of carotenogenic genes
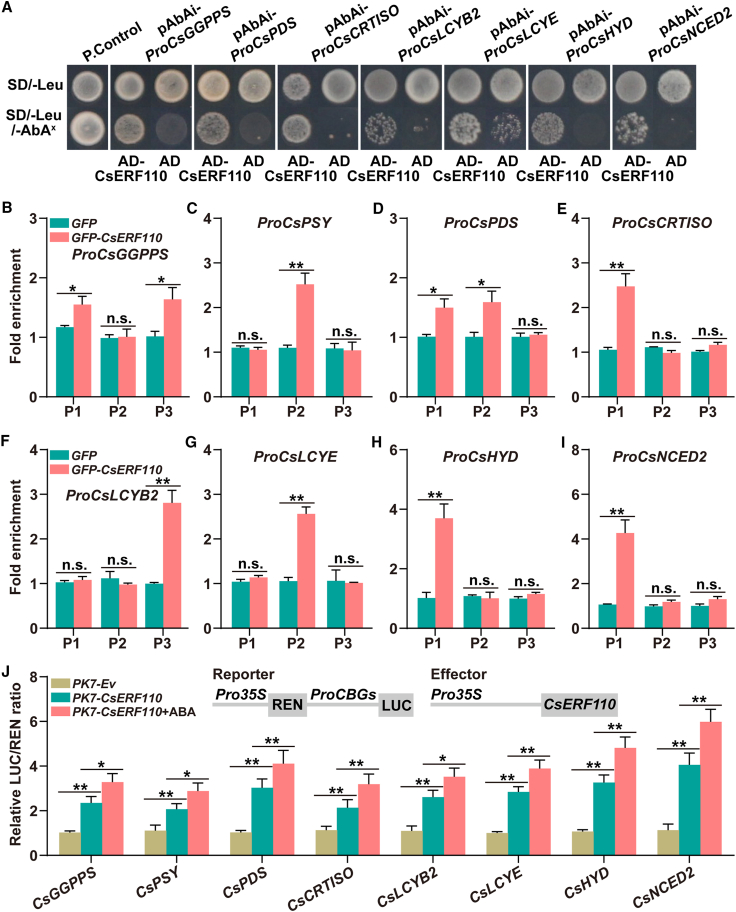
The transgenic experiment identified the carotenogenic target genes of CsERF110 (CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, and CsNCED2) (Figure 1H and 1L). To clarify the regulatory role of CsERF110 with respect to these target genes, we performed yeast one-hybrid (Y1H) assays. All strains grew well on SD/-Leu selective medium, but only the strains transformed with CsERF110 and the promoters of the above target genes, as well as the positive controls, remained alive on SD/-Leu/Aureobasidin A (AbA)x selective medium (Figure 2A). These results indicate that CsERF110 binds directly to the promoters of CsGGPPS, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, and CsNCED2.
Figure 2.
CsERF110 directly regulates the transcription of carotenogenic genes.
(A) A Y1H assay identified interactions of CsERF110 with the promoters of carotenogenic genes. PGADT7-Rec-p53+p53-AbAi and empty PGADT7+pAbAi-ProCBGs served as positive (P. Control) and negative (N. Control) controls, respectively. Aureobasidin A was the yeast cell growth inhibitor. SD/-Leu medium was supplemented with 200 ng mL−1 Aureobasidin A (ProCsGGPPS, ProCsPDS, ProCsLCYB2, and ProCsLCYE) or 150 ng mL−1 Aureobasidin A (ProCsCRTISO, ProCsHYD, and ProNCED2).
(B–I) ChIP–PCR assays showed the interaction of CsERF110 with several regions in the promoters of carotenogenic genes. Cross-linked chromatin samples were extracted from GFP-CsERF110 fruit calli and precipitated with an anti-GFP antibody. The eluted DNA fragment was amplified by qPCR.
(J) Dual-luciferase assays indicated that CsERF110 enhances the transcription of carotenogenic genes, and this enhancement is strengthened by ABA treatment. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination. Data are presented as means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).
We next searched for potential CsERF110 binding motifs on these target gene promoters and found at least one putative motif on each (Supplemental Figure 3). To define the specific CsERF110 binding motifs on the target gene promoters, CsERF110 was cloned into the GFP-tag vector. The resulting GFP-CsERF110 fusion vector was then stably transformed into citrus calli, and citrus calli transformed with empty GFP served as the negative controls. Positive transgenic lines were selected and used for a chromatin immunoprecipitation (ChIP) PCR assay. The ChIP–PCR results showed that the CsERF110 signal was enriched on at least one potential binding motif in each target gene promoter (Figure 2B–2I). These results further confirmed that CsERF110 binds directly to the promoters of its target genes in vivo.
To determine the effect of CsERF110 on the activity of carotenoid biosynthesis gene (CBG) promoters, the promoters of CBGs were cloned into pGreen0800-II as reporters, and PK7-CsERF110 was used as the effector. The resulting reporters and effectors were transiently co-transformed into N. benthamiana leaves. The LUC results showed that overexpression of CsERF110 significantly increased the LUC/REN ratio of the CBG promoters, which was further enhanced by ABA treatment (Figure 2J). These results indicated that ABA-induced CsERF110 binds directly to the promoters of downstream carotenoid metabolism target genes, thus activating their expression.
CsERF110 interacts with CsERF53 to activate CsERF53 expression
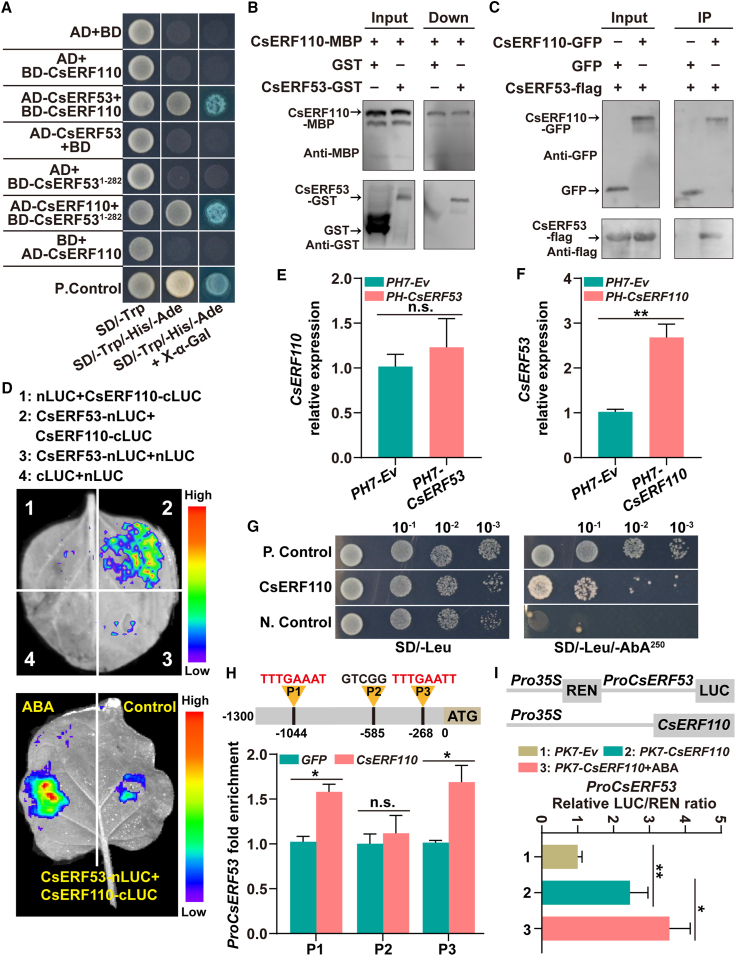
Many ERFs form homodimers or heterodimers with their interacting proteins to participate in the regulation of various biological processes (Sasaki et al., 2007; Kim et al., 2018; Xie et al., 2019; Zhang et al., 2023). To identify proteins that interact with CsERF110, we performed Y2H screening with CsERF110 as the prey, and the results showed no CsERF110 transcriptional self-activation in yeast cells (Supplemental Figure 4). After multiple Y2H screenings, one gene (orange1.1t00851) encoding an ERF protein was screened multiple times. Therefore, orange1.1t00851 was subsequently analyzed as a potential protein interacting with CsERF110. Phylogenetic analysis showed that orange1.1t00851 was closely clustered with the ERF protein AtERF53 (AT2G20880) from A. thaliana, and orange1.1t00851 was therefore renamed CsERF53 (Supplemental Figure 5). A transcriptional self-activation analysis showed that full-length CsERF53 exhibited self-activation, but this activation disappeared after truncation at position 282 of the amino acid sequence (Supplemental Figure 6). On the basis of this result, we constructed AD-CsERF53 and BD-ERF511−282 fusion yeast vectors to verify the interaction between CsERF53 and CsERF110. All recombinant strains grew well on SD/-Trp/-Leu medium, and BD-CsERF110+AD-CsERF53, BD-CsERF531−282+AD-CsERF110, and the positive control remained alive on SD/-Trp/-Leu/-His/-Ade with X-α-Gal (Figure 3A), verifying the in vivo protein interaction between CsERF53 and CsERF110.
Figure 3.
CsERF110 binds CsERF53 and transcriptionally activates CsERF53.
(A) A Y2H assay reveals an interaction between CsERF53 and CsERF110. Yeast grown in SD/-Trp/-Leu medium and SD/-Trp/-Leu/-His/-Ade medium are shown. The interaction is indicated by yeast growth and X-α-Gal staining.
(B) The interaction between CsERF53 and CsERF110 was analyzed using a pull-down assay. The band detected by the GST antibody in the pull-down protein sample indicates the interaction between CsERF53 and CsERF110.
(C) The interaction between CsERF53 and CsERF110 was confirmed with a coIP assay. GFP antibody beads were used for IP. Anti-GFP and anti-FLAG antibodies were used for immunoblot analyses. The band detected by the GFP antibody in the IP samples indicates an interaction between CsERF53 and CsERF110.
(D) A luciferase complementation imaging assay shows that CsERF53 interacts with CsERF110, and this interaction is enhanced by ABA treatment. Luciferase activity was imaged in the indicated regions 3 days after infiltration. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination.
(E and F) Expression of CsERF110(E) and CsERF53(F) in transgenic calli.
(G) A Y1H assay identified an interaction of CsERF110 with the CsERF53 promoter. PGADT7-Rec-p53+p53-AbAi and empty PGADT7+pAbAi-ProCsERF53 served as the positive (P. Control) and negative (N. Control) controls, respectively. Aureobasidin A was the yeast cell growth inhibitor.
(H) ChIP–PCR assays show the interaction of CsERF110 with several regions in the promoter of CsERF53. The black lines represent the putative binding motifs of ERF family proteins in the CsERF53 promoter. Cross-linked chromatin samples were extracted from GFP-CsERF110 fruit calli and precipitated with an anti-GFP antibody. The eluted DNA fragment was amplified by qPCR.
(I) Dual-luciferase assays indicated that CsERF110 positively regulates CsERF53 transcription, and this effect is enhanced by ABA treatment. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination. Data are presented as means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).
To further confirm the in vivo interaction between CsERF53 and CsERF110, CsERF53-GST (glutathione S-transferase) and CsERF110-MBP (maltose-binding protein) fusion proteins were prepared for use in a pull-down assay. The results showed that GST-tagged CsERF53 interacted with MBP-tagged CsERF110 in vitro (Figure 3B). Next, FLAG-CsERF110 was co-transformed into N. benthamiana leaves with GFP-CsERF53 or the GFP empty vector for coIP assays. As shown in Figure 3C, GFP-CsERF53, GFP, and FLAG-ERF110 were all present in the input groups, whereas FLAG-CsERF110 was present only in the FLAG-CsERF110+GFP-CsERF53 groups. These results further confirmed that CsERF53 in vivo interacts with CsERF110 in plants. Subsequently, luciferase complementation imaging assays revealed a strong fluorescence signal in the region co-expressing CsERF53-nLUC and CsERF110-cLUC (Figure 3D), and this fluorescence signal was further enhanced by ABA treatment (Figure 3D), indicating that CsERF53 physically interacts with the CsERF110 protein in vivo.
To further explore the relationship between CsERF110 and CsERF53, we detected the expression of CsERF110 and CsERF53 in CsERF53-overexpressing and CsERF110-overexpressing calli, respectively. Quantitative real-time PCR results showed that there was no significant difference in CsERF110 expression between CsERF53-overexpressing calli and controls (Figure 3E), but the CsERF53 mRNA level was significantly higher in CsERF110-overexpressing calli than in the controls (Figure 3F). On the basis of this result, we speculated that CsERF53 might be a target gene of CsERF110. To verify this speculation, we analyzed the promoter of CsERF53 using PLACE and identified three potential cis-elements of CsERF110 (Supplemental Figure 7). Y1H and ChIP–PCR experiments showed that CsERF110 directly bound to the CsERF53 promoter in vivo (Figure 3G and 3H). A LUC activity assay indicated that the CsERF53 promoter was activated by CsERF110 and that this activation was enhanced by ABA treatment (Figure 3I). These data demonstrated that CsERF110 interacts with CsERF53 and activates its expression and that ABA signaling enhances their interaction and regulation.
CsERF53 participates in ABA-induced carotenoid accumulation as an essential nuclear-localized transcriptional activator
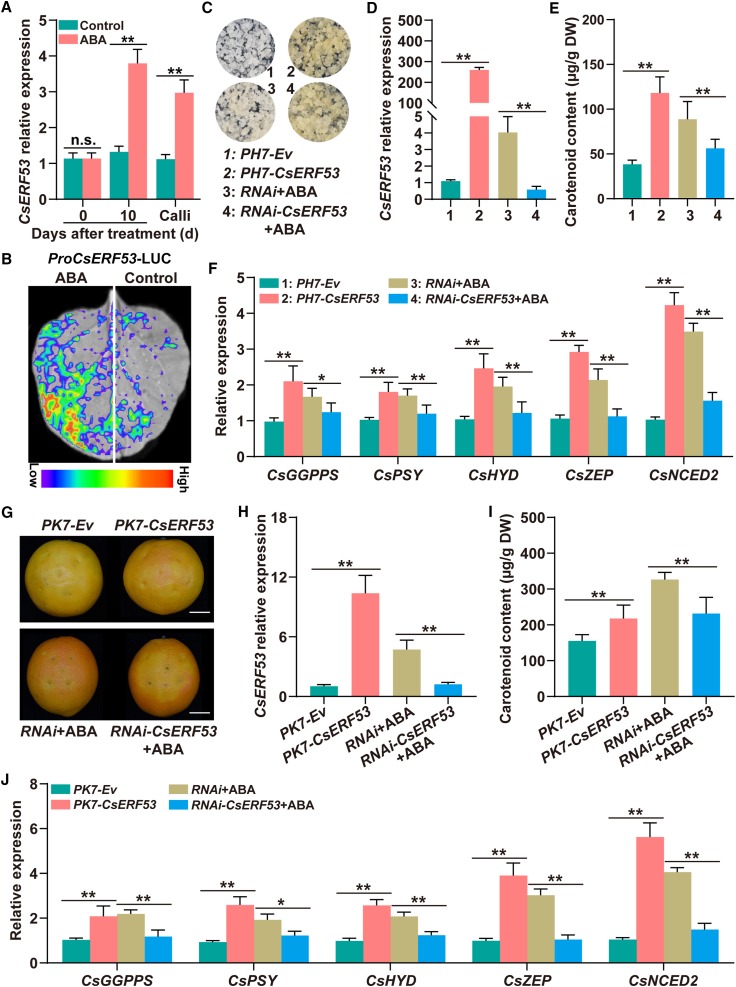
To clarify the role of CsERF53 in ABA-induced carotenoid accumulation, we examined the expression of CsERF53 in ABA-treated citrus fruit and calli. Quantitative real-time PCR results showed that ABA treatment promoted fruit and calli coloration and also upregulated CsERF53 expression (Figure 4A). In addition, ProCsERF110-LUC produced higher fluorescence intensity in ABA-treated regions than in water-treated regions (Figure 4B). These results indicate that CsERF53 is induced by ABA signaling and may be involved in regulating ABA-mediated carotenoid accumulation in citrus. We next analyzed the subcellular localization and transcriptional activity of CsERF53. A subcellular localization assay showed that CsERF53 was localized in the nucleus (Supplemental Figure 8A). Yeast experiments suggested that CsERF53 exhibited transcriptional activity (Supplemental Figure 8B). Transient transformation results indicated that CsERF53 had transcriptional activation activity in N. benthamiana leaves (Supplemental Figure 8C and 8D). Together, these results demonstrated that CsERF53 serves as a nuclear-localized transcriptional activator and is induced by ABA signaling, thus potentially participating in ABA-mediated citrus carotenoid biosynthesis.
Figure 4.
CsERF53 is essential for ABA-mediated carotenoid biosynthesis in citrus.
(A)CsERF110 expression in citrus fruit and calli under ABA treatment.
(B)In vivo LUC complementation imaging of N. benthamiana leaves treated with water or ABA. A suspension of Agrobacterium GV3101 carrying ProCsERF53-LUC was injected into N. benthamiana leaves. Two days after injection, N. benthamiana leaves were treated with water or 100 μM ABA, and 24 h later, bioluminescence imaging was used to measure the luciferase activity of ProCsERF53.
(C–F) Stable transformation of CsERF53 in citrus calli. (C) Phenotypes. PH7-CsERF53 and RNAi-ERF53 indicate CsERF53 overexpression and RNA interference, respectively. PH7-Ev and RNAi are controls. Expression levels of CsERF53(D) and CsGGPPS, CsPSY, CsZEP, CsHYD, and CsNCED2(F) are shown. (E) Total carotenoid content (μg/g DW).
(G, H, and J) Transient expression of CsERF53 in citrus fruit. (G) Phenotypes. PK7-Ev and RNAi serve as controls. PK7-CsERF53 and RNAi-CsERF53 indicate CsERF53 overexpression and RNA interference, respectively. Scale bars, 2 cm. Transcript levels of CsERF53(H) and CsGGPPS, CsPSY, CsZEP, CsHYD, and CsNCED2(J) are shown.
(I) Total carotenoid content (μg/g DW). Data are presented as means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).
To further investigate the function of CsERF53 in ABA-induced citrus carotenoid biosynthesis, we performed stable transformation and transient transformation experiments in citrus calli and fruits, respectively (Figure 4C–4J). Citrus fruits and calli overexpressing CsERF53 displayed more coloration than controls (Figure 4C, 4D, 4G, and 4H). Accordingly, HPLC results showed that the carotenoid content was significantly higher in CsERF53-overexpressing calli and fruits than in the controls (Figure 4E and 4I). Quantitative real-time PCR revealed that expression of five carotenoid metabolism genes, CsGGPPS, CsPSY, CsHYD, CsZEP, and CsNCED2, was significantly upregulated in CsERF53-overexpressing calli and fruits compared with controls (Figure 4F and 4J). These results indicate that CsERF53 positively regulates carotenoid accumulation in citrus by activating the expression of multiple CBGs.
We also treated CsERF53-RNAi calli and fruits with an ABA solution. ABA treatment significantly promoted the coloration of fruits and calli in the control group, but it had no obvious effect on CsERF53-RNAi calli and citrus fruits (Figure 4C, 4D, 4G, and 4H). Similarly, the ABA-treated control group exhibited higher carotenoid content than CsERF53-RNAi calli and fruits (Figure 4E and 4I). In addition, after ABA treatment, mRNA levels of CsGGPPS, CsPSY, CsHYD, CsZEP, and CsNCED2 were significantly higher in the control group than in CsERF53-RNAi calli and fruits (Figure 4F and 4J). Together, these results confirmed that CsERF53 participates in ABA-induced carotenoid accumulation in citrus as an essential activator by activating the expression of several carotenoid biosynthetic genes.
ABA signaling enhances the CsERF53-activated transcriptional regulation of carotenogenic genes
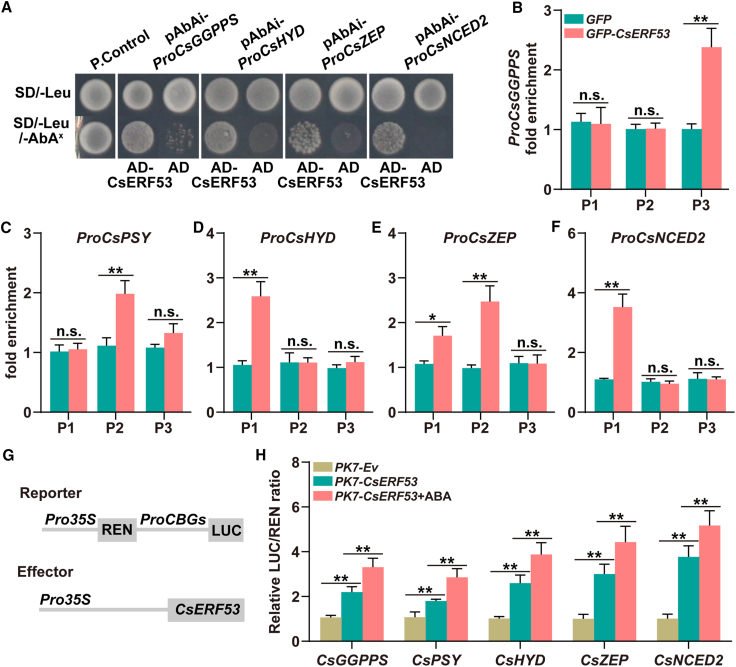
We first performed Y1H experiments to clarify the regulatory effects of CsERF53 on the carotenogenic genes CsGGPPS, CsHYD, CsZEP, and CsNCED2. As shown in Figure 5A, all strains grew well in SD/-Leu selective medium, but only the AD-ERF53-containing strain and the positive control remained alive in SD/-Leu/AbAx medium, suggesting an interaction between CsERF53 and the promoters of carotenogenic genes. The DNA fragments of potential CsERF53-binding elements were amplified from GFP-CsERF53 transgenic calli, and enrichment of the CsERF53 signal was examined. The ChIP–PCR results showed that CsERF53 signals were significantly enriched on at least one potential motif of the CsPSY, CsGGPPS, CsHYD, CsZEP, and CsNCED2 promoters (Figure 5B–5F andSupplemental Figure 9), suggesting that CsERF53 directly binds to the promoters of CsPSY, CsGGPPS, CsHYD, CsZEP, and CsNCED2 in vivo.
Figure 5.
CsERF53 directly regulates the transcription of carotenogenic genes.
(A) A Y1H assay identified interactions of CsERF53 with the promoters of carotenogenic genes. PGADT7-Rec-p53+p53-AbAi and empty PGADT7+pAbAi-ProCBGs served as positive (P. Control) and negative (N. Control) controls, respectively. Aureobasidin A was the yeast cell growth inhibitor. SD/-Leu medium was supplemented with 200 ng mL−1 Aureobasidin A (ProCsGGPPS and ProCsZEP) or 150 ng mL−1 Aureobasidin A (ProCsHYD and ProNCED2).
(B–F) ChIP–PCR assays showed the interaction of CsERF53 with several regions in the promoters of carotenogenic genes. Cross-linked chromatin samples were extracted from GFP-CsERF53 fruit calli and precipitated with an anti-GFP antibody. The eluted DNA fragment was amplified by qPCR.
(G) Schematic representation of reporter and effector constructs used in the dual-luciferase assays.
(H) Dual-luciferase assays indicated that CsERF53 enhances the transcription of carotenogenic genes. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination. Data are presented as means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).
We also performed dual-luciferase assays to investigate the regulatory effects of CsERF53 on the promoter activity of the downstream target genes CsPSY, CsGGPPS, CsHYD, CsZEP, and CsNCED2 (Figure 5G). The relative LUC/REN ratio was significantly higher in PK7-CsERF53 than in controls, and the activation effect of CsERF53 on these target genes was further enhanced by ABA treatment (Figure 5H). All the above results indicate that CsERF53 binds directly to the promoters of CsPSY, CsGGPPS, CsHYD, CsZEP, and CsNCED2 and activates their expression, an effect that is further enhanced by ABA signaling.
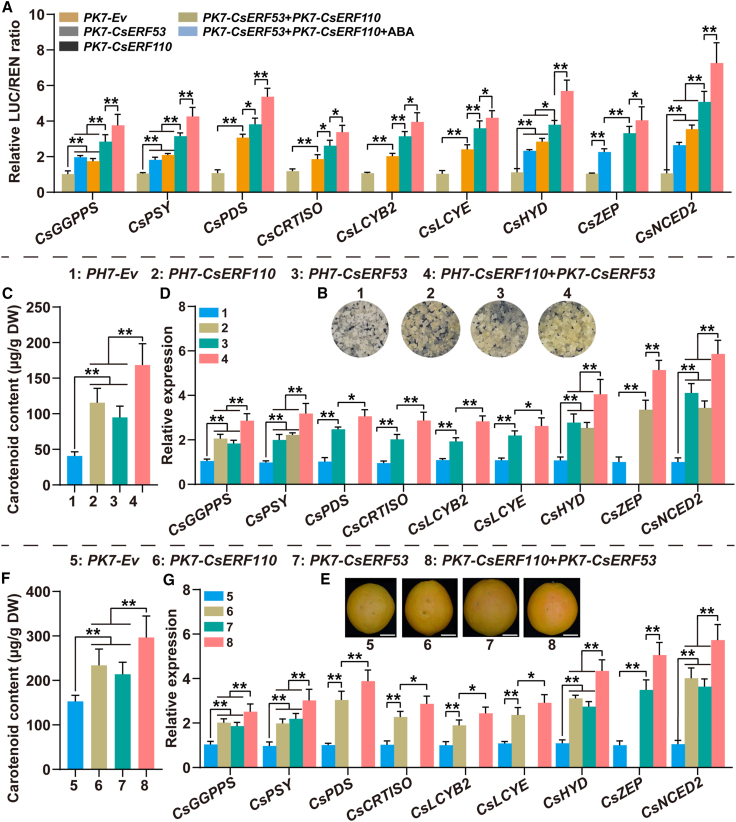
CsERF110 and CsERF53 synergistically regulate carotenoid accumulation in citrus
As demonstrated above, both CsERF53 and CsERF110 are involved in regulating ABA-mediated citrus carotenoid biosynthesis as positive regulators (Figures 1, 2, 4, and 5). CsERF110 not only formed a protein complex with CsERF53 but also bound directly to the CsERF53 promoter to activate CsERF53 expression, thus forming the transcriptional regulatory module CsERF110–CsERF53 (Figure 3). To clarify the regulatory effect of this module on carotenoid metabolism in citrus, we performed dual-luciferase assays. As shown in Figure 6A, compared with overexpression of CsERF110 alone or CsERF53 alone, simultaneous overexpression of CsERF110 and CsERF53 more significantly enhanced the transcriptional activation effects on downstream target genes, and these effects were further enhanced by ABA treatment (Figure 6A). These results indicate that the transcriptional regulatory module CsERF110–CsERF53 positively regulates downstream target genes.
Figure 6.
The transcriptional regulatory module CsERF110–CsERF53 positively regulates carotenoid biosynthesis in citrus.
(A) Effect of the transcriptional regulatory module CsERF110–CsERF53 on the regulation of the promoter activity of downstream target genes. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination.
(B–D) Stable transformation of CsERF110 and CsERF53 in citrus calli. PH7-Ev is the control. PH7-CsERF110/CsERF53 indicates overexpression of CsERF110 or CsERF53. PH7-CsERF110+PK7-CsERF53 indicates co-overexpression of CsERF110 and CsERF53. (B) Phenotypes. (C) Carotenoid content (μg/g DW). (D) The expression levels of CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, CsZEP, and CsNCED2.
(E–G) Transient expression of CsERF110 and CsERF53 in citrus fruit. PK7-Ev is the control. PK7-CsERF110/CsERF53 indicates overexpression of CsERF110 or CsERF53. PK7-CsERF110+PK7-CsERF53 indicates co-overexpression of CsERF110 and CsERF53. Scale bars, 3 cm. (E) Phenotypes. (F) Carotenoid content (μg/g DW). (G) Expression levels of CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, CsZEP, and CsNCED2. Data represent means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).
We further verified the role of the CsERF110–CsERF53 module in regulating carotenoid metabolism in citrus calli and fruits. Overexpression of CsERF110 alone or CsERF53 alone promoted coloration in citrus calli and fruit (Figure 6B and 6E) and increased their carotenoid content (Figure 6C and 6F) by activating carotenogenic gene expression (Figure 6D and 6G). More importantly, simultaneous overexpression of CsERF110 and CsERF53 further enhanced their positive regulatory effects on carotenoid accumulation in citrus (Figure 6B–6G). All these results indicate that CsERF110 and CsERF53 form a CsERF110–CsERF53 transcriptional regulatory module, thus positively regulating carotenoid accumulation in citrus.
Discussion
The findings of the present study reveal that the ABA-induced transcriptional regulatory module CsERF110–CsERF53 links ABA signaling to carotenoid accumulation, thus promoting citrus fruit coloration.
In multiple studies, ABA has been identified as the main inducer of the ripening process in non-climacteric fruit (Li et al., 2022a, 2022b; Wang et al., 2022; Sun et al., 2024). In addition to the phytohormone ABA, ETH has been identified as an important effector in the non-climacteric fruit-ripening process (Merchante et al., 2013; Hou et al., 2018; Zhu et al., 2021). ABA plays important roles in ETH-dominated ripening of climacteric fruit such as pear, peach, tomato, apple, and banana, where ABA is mainly involved in the fruit development and coloration process (Dai et al., 2014; Wang et al., 2019; Seale, 2022; Zou et al., 2022; Jia et al., 2024). On the basis of these findings, we speculate that there may be crosstalk between ABA and ETH during the regulation of fruit ripening and senescence in both climacteric and non-climacteric fruits. Our data show that both CsERF53 and CsERF110 are induced by ABA signaling to participate in ABA-promoted carotenoid accumulation in citrus (Figures 1 and 4). Unlike CsERF53 and CsERF110, CsERF061 is induced by ETH and positively regulates citrus carotenoid accumulation (Zhu et al., 2021). In addition, overexpression of CsERF53, CsERF110, or CsERF061 can promote the accumulation of ABA in citrus (Supplemental Figure 10; Zhu et al., 2021). These results indicate that although these three ERFs (CsERF53, CsERF110, and CsERF061) are activated by different hormone signals (ABA or ETH), they all participate in regulating citrus carotenoid metabolism, implying some type of regulatory or functional relationship among CsERF53, CsERF110, and CsERF061 that will need to be explored further in future studies.
ERFs, as well-known downstream ETH signaling components, play important roles in ETH-dependent developmental processes such as softening, flowering, coloration, and ripening in climacteric fruits, and most ERFs are directly responsive to and transduce ETH signaling (Li et al., 2017; Gao et al., 2020; Wu et al., 2021; Deng et al., 2022). However, numerous studies have shown that ERFs are also involved in the ripening process of non-climacteric fruits such as strawberry, loquat, grape, and pepper (Zhang et al., 2018, 2020; Jia et al., 2023; Song et al., 2023; Xu et al., 2023). The ERFs of citrus, a typical non-climacteric fruit, regulate a variety of biological processes. The interaction between CitERF13 and CitVHA greatly enhances citrus fruit flavor by promoting citric acid accumulation (Li et al., 2016). CitERF13 accelerates chlorophyll degradation in citrus by activating CitPPH expression (Yin et al., 2016). Three AP2/ERF TFs (CitRAV1, CitERF32, and CitERF33) positively modulate flavonoid biosynthesis in citrus (Zhao et al., 2021). ETH-induced CsERF061 positively regulates carotenoid accumulation by activating the expression of nine key carotenoid pathway genes (Zhu et al., 2021). As a downstream factor of ETH signaling, PtrERF9 improves the cold resistance of Poncirus trifoliata by regulating reactive oxygen species homeostasis in response to cold stress (Zhang et al., 2022). CiERF023 and CiERF3 cooperatively regulate CiGA20ox1/2-mediated citrus plant height (Chu et al., 2023). A regulatory module ERF TF, CsDRNL, activates MYC5, thus facilitating the development of oil glands in citrus (Wang et al., 2024). In our study, we found that the promoters of CsERF110 and CsERF53 responded significantly to ABA signaling and that ABA treatment significantly promoted the expression of CsERF110 and CsERF53 (Figures 1C, 1D, 4A, and 4B). Our transgenic analysis revealed that CsERF110 and CsERF53 play essential roles in ABA-induced carotenoid accumulation (Figures 1E–1L and 4C–4J). Molecular biochemical data demonstrated that CsERF110 and CsERF53 bind directly to the promoters of multiple carotenoid metabolism genes and upregulate their expression, ultimately positively regulating carotenoid accumulation (Figures 2 and 5). As far as we know, this is the first time that ABA-induced ERF TFs have been reported to regulate carotenoid metabolism during non-climacteric fruit ripening.
The ERF TFs in apple, a typical climacteric fruit, can respond to two hormone signals (ETH and ABA) and participate in the regulation of pigment (anthocyanin and carotenoid) metabolism during apple fruit ripening (An et al., 2018; Jia et al., 2024). Similarly, this study and our previous study found that ETH-induced CsERF061 and ABA-induced CsERF53 and CsERF110 are involved in regulating carotenoid biosynthesis in citrus (Figures 1E–1L and 4C–4J; Zhu et al., 2021). These findings imply that ERF TFs are involved in fruit ripening and senescence in both climacteric and non-climacteric fruits. The present study deepens our understanding of this property of ERF TFs.
Our study also revealed a protein–protein interaction between CsERF53 and CsERF110, which enhanced their regulatory effect on carotenoid metabolism (Figures 3A–3D and 6A). ERF TFs have been extensively reported to form heterodimers with other types of proteins to participate in the regulation of various biological processes (Deng et al., 2022; Li et al., 2022a, 2022b; Zhang et al., 2022; Chu et al., 2023; Jia et al., 2023). To date, there have been few studies on homodimers or heterodimers formed by two ERF TFs (Huang et al., 2021; Li et al., 2022; Zhang et al., 2023), especially with regard to their regulatory effects on carotenoid metabolism. Our work is the first to report the protein complex composed of two ERFs, CsERF53 and CsERF110, and their regulatory effects on carotenoid accumulation, enriching our understanding of the functional diversity of ERF TFs and the molecular regulatory mechanisms of plant carotenoid metabolism. In addition to forming a heterodimer with CsERF53, CsERF110 can also bind directly to the CsERF53 promoter and activate CsERF53 expression to form a transcriptional cascade (Figure 3E–3I), further synergistically regulating carotenoid metabolism.
Feedback regulation is a complex and finely tuned control mechanism that acts in a wide array of plant developmental processes. In ABA signal transduction, AtABI5 directly activates the expression of the ABA receptor AtPYL11/12, further elevating ABA levels and inducing AtABI5, eventually forming a positive feedback loop to regulate ABA-mediated seed germination (Zhao et al., 2020). Drought stress induces MdMYB88 and MdMYB124 to positively regulate ABA accumulation to resist drought stress, and in return, ABA represses the expression of both genes, thus avoiding excessive defense against drought (Xie et al., 2021). In the carotenoid metabolism process, CsMPK6 interacts with CsMYC2 to form a jasmonate-induced CsMPK6–CsMYC2 negative feedback loop, further regulating β-citraurin accumulation and fruit coloration in citrus (Yue et al., 2023). Furthermore, carotenoid metabolism is the only source of precursors for ABA biosynthesis, and the NCEDs responsible for carotenoid metabolism are the key rate-limiting enzymes of ABA biosynthesis (Chen et al., 2020). Hence, carotenoid biosynthesis and ABA biosynthesis are closely linked. In the present study, both ABA and carotenoid levels were significantly increased in calli and fruits overexpressing CsERF110 and CsERF53 (Figures 1G, 1K, 4E, and 4I; Supplemental Figure 10). On the one hand, CsERF110 and CsERF53 directly activate the expression of CsNCED2 to promote ABA biosynthesis (Figures 2 and 5). On the other hand, CsERF110 and CsERF53 directly induce multiple carotenoid biosynthetic genes, thus promoting carotenoid biosynthesis and providing sufficient precursors for ABA biosynthesis, eventually facilitating ABA accumulation (Figures 1, 2, 4, and 5). In addition, CsERF110 and CsERF53 can respond to ABA signaling and are significantly induced by ABA (Figures 1C, 1D, 4A, and 4B). Therefore, there is a positive feedback loop between ABA biosynthesis and carotenoid metabolism, and this positive feedback loop is very important for ABA-mediated fruit ripening and coloration in citrus. CsERF110 and CsERF53, as core components of the feedback loop, play essential roles in ABA-induced fruit coloration and carotenoid accumulation, making the CsERF110–CsERF53 module the hub of the citrus fruit-ripening process.
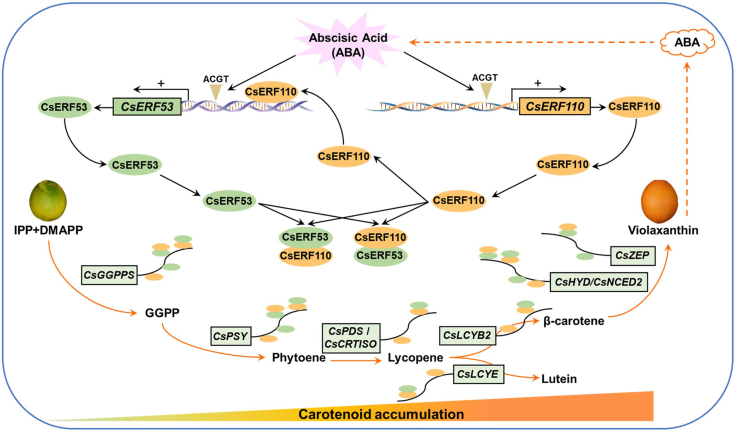
On the basis of these findings, we propose a working model in which the CsERF110–CsERF53 transcriptional regulatory module mediates a positive feedback regulatory loop between ABA and carotenoid metabolism (Figure 7). Our data show that CsERF110 responds to ABA signaling and activates the expression of multiple carotenogenic genes (CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, and CsNCED2) to promote carotenoid accumulation. Meanwhile, CsERF53 is not only induced by ABA signaling but also directly activated by CsERF110, in turn activating the expression of several carotenogenic genes (CsGGPPS, CsPSY, CsHYD, and CsNCED2) and ultimately promoting carotenoid biosynthesis. CsERF110 also interacts with CsERF53, further activating their downstream carotenogenic genes. In return, ABA, as ripening signal, significantly induces the expression of CsERF110 and CsERF53 and their interaction through feedback regulation, thereby promoting carotenoid biosynthesis and fruit coloration. Taken together, our results demonstrate a positive feedback regulatory loop between the CsERF110–CsERF53 regulatory module and ABA signaling that determines carotenoid biosynthesis in citrus.
Figure 7.
Model for the positive feedback regulatory loop between ABA metabolism and carotenoid metabolism mediated by the transcriptional regulatory module CsERF110–CsERF53 in citrus.
CsERF110 responds to ABA signaling and activates the expression of multiple carotenogenic genes (CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, and CsNCED2) to promote carotenoid accumulation. Meanwhile, CsERF53 is not only induced by ABA signaling but also directly activated by ERF110, which in turn activates the expression of several carotenogenic genes (CsGGPPS, CsPSY, CsHYD, and CsNCED2), ultimately promoting carotenoid biosynthesis. In addition, CsERF110 interacts with CsERF53, and their interaction further enhances their activation of downstream carotenogenic genes. In turn, ABA, as a ripening signal, significantly induces the regulation and interaction of CsERF110 and CsERF53 in feedback regulation, thereby promoting carotenoid biosynthesis and fruit coloration. In general, a positive feedback regulatory loop between the CsERF110–CsERF53 regulatory module and ABA signaling determines carotenoid biosynthesis in citrus.
This study provides new insights into the regulation of fruit ripening and senescence by ERFs in non-climacteric fruits. Our findings reveal the functional diversity of ERFs and the molecular mechanism of ripening in non-climacteric and climacteric fruit. Considering the importance of carotenoid content for the nutritional and aesthetic value of citrus and many other carotenoid-rich fruit crops, characterization of the molecular mechanisms that underlie ABA-mediated carotenoid biosynthesis in plants will facilitate the development of transgenic/gene-editing approaches, ultimately contributing to the quality improvement of citrus and other fruit crops.
Materials and methods
Plant materials and treatments
‘Valencia’ orange fruits (Citrus sinensis Osbeck) were used in this study. Citrus calli were subcultured at 20-day intervals on solid Murashige Tucker (MT) medium.
Citrus fruits were picked at 190 days after flowering and treated with 500 μM ABA solution (Sigma A1049) or with sterile water as a control. All fruits were naturally dried at room temperature and then placed in a dark phytotron at 25°C. Peels were sampled along the equatorial plane of fruits. Citrus calli were subcultured on MT medium containing 250 μM ABA or sterile water (as a control). All samples (calli and peels) were immediately frozen in liquid nitrogen and stored at −80°C until further analysis.
ABA production
Extraction and measurement of endogenous ABA were performed as described previously (Ma et al., 2014; He et al., 2018; Sun et al., 2023). At least three biological replicates from independent extractions were performed.
Quantitative real-time PCR
Quantitative real-time PCR was performed as reported previously (Zhang et al., 2021; Sun et al., 2024). For citrus fruit, the peels from three independent groups were used as three replications; for citrus calli, each line of transgenic calli was used as one biological replicate. Three replicates were used in each experiment. Each replicate was performed in triplicate. The quantitative real-time PCR data were analyzed using the E−ΔΔCt method. Two ACTIN genes served as internal controls in the quantitative real-time PCR assays. The primers used for quantitative real-time PCR are listed in Supplemental Table 1.
Determination of carotenoid content
See the supplemental information for details.
Plasmid construction, genetic transformation, and transient expression
The coding sequences (CDSs) of CsERF110 and CsERF53 were cloned into the PK7WG2D (PK7) and PH7WG2D (PH7) vectors to construct the overexpression vectors PH7/PK7-CsERF110/CsERF53. The RNA interference vectors RNAi-CsERF110/CsERF53 were generated by cloning the truncated CDS (1–303) of CsERF110 and the truncated CDS (1–273) of CsERF53 into pRI101 (RNAi). Stable transformation of citrus calli and transient injection of citrus fruit were performed as described previously (Sun et al., 2023).
Gene cloning and phylogenetic analysis
The CDS and promoter sequences of CsERF53 and CsERF110 were amplified according to the reference genome CPBD (http://citrus.hzau.edu.cn/). A phylogenetic tree was constructed using MEGA 7.0. All primers are listed in Supplemental Table 1.
Y2H assay
See the supplemental information for details.
Subcellular localization assay
See the supplemental information for details.
Pull-down and coIP assays
The CDSs of CsERF110 and CsERF53 were separately cloned into pMAL-C2X and pGEX4T-1 for expression of fusion proteins (CsERF110-MBP and CsERF53-GST). The CsERF110-MBP fusion protein was purified with MBP magnetic beads and incubated with CsERF53-GST for 2 h on ice. GST protein was used as the negative control. The MBP-tagged protein was eluted using elution buffer containing maltose, and the eluted proteins were analyzed by immunoblotting with anti-GST and anti-MBP antibodies.
Two fusion constructs, CsERF110-GFP and CsERF53-FLAG, were co-expressed in N. benthamiana leaves by A. tumefaciens-mediated transformation. The empty GFP vector and CsERF53-FLAG were co-expressed as a negative control. After a 3-day incubation, leaf powder was added to protein extraction buffer (50 mM Tris–MES [pH 8.0], 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA [pH 8.0], 5 mM DTT, and 1 mM PMSF), shaken on ice for 30 min, and centrifuged for 30 min at 12 000 rpm. The supernatant was incubated with anti-GFP magnetic beads to immunoprecipitate either CsERF110-GFP or GFP. The immunoprecipitate was examined by immunoblotting with an anti-FLAG antibody.
Y1H assay
The promoters of target genes were cloned into the pAbAi vector to obtain the baits (pAbAi-ProCsCBGs and pAbAi-ProCsERF53). The CDSs of CsERF110 and CsERF53 were cloned into PGADT7 vector as prey. The prey and bait constructs were co-transformed into yeast Y1H Gold according to the manufacturer’s instructions (TaKaRa). PGADT7-Rec-p53+p53-AbAi and PGADT7+baits served as the positive control and negative control, respectively.
Transcriptional activation analysis
See supplemental information for details.
ChIP–PCR analysis
Transgenic 35Spro::CsERF110-GFP, 35Spro::CsERF53-GFP, and 35Spro::GFP citrus calli were used for ChIP_PCR analysis with an anti-GFP antibody as described by Sun et al. (2024). Three transgenic lines of citrus calli were used for ChIP–PCR analysis with three biological replicates, and each biological replicate was repeated three times. The primers used for ChIP–PCR are listed in Supplemental Table 1.
Dual luciferase reporter assay
See supplemental information for details.
Luciferase complementation assay
The CDSs of CsERF110 and CsERF53 were separately cloned into the JW-772-cLUC and JW-771-nLUC vectors and then co-overexpressed in N. benthamiana leaves via A. tumefaciens GV3101-mediated transformation. nLUC+CsERF110-cLUC, CsERF53-nLUC+cLUC, and nLUC+cLUC served as controls. Luciferase activity was measured using a NightSHADE LB 985 imaging system (Berthold Technologies, Germany) following a 3-day incubation.
Statistical analyses
The statistical analysis of data was performed using Microsoft Office (2010) and GraphPad 8.0 software. The data are presented as means ± SDs of three biological replicates. Asterisks indicate statistically significant differences as determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).
Accession numbers
Sequence data from this study can be found in the reference genome of CPBD (http://citrus.hzau.edu.cn/). All accession numbers for this study are listed in Supplemental Table 1.
Funding
This work was supported by the National Key R&D Program of China (2023YFD2300600), the National Natural Science Foundation of China (no. 31930095), and the National Modern Agricultural (Citrus) Technology Systems of China (no. CARS-27).
Acknowledgments
All authors have no conflict of interest to declare.
Author contributions
X.D. supervised the research; Q.S. and X.D. designed the experiments; Q.S. performed the experiments, with contributions from Z.H., D.F., R.W., and Y.Z.; Q.S. and Z.H. wrote the manuscript; X.D. and Q.S. revised the manuscript; J.Y., L.C., J.X., Y.C., and Q.X. provided critical comments on the manuscript editing.
Published: August 20, 2024
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- An J.P., Wang X.F., Li Y.Y., Song L.Q., Zhao L.L., You C.X., Hao Y.J. EIN3-like1, MYB1, and ethylen response factor 3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018;178:808–823. doi: 10.1104/pp.18.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L.L., Yan Z., Sheng X.X., Liu H.Q., Wang Q.Y., Zeng R.F., Hu C.G., Zhang J.Z. Citrus ACC synthase CiACS4 regulates plant height by inhibiting gibberellin biosynthesis. Plant Physiol. 2023;192:1947–1968. doi: 10.1093/plphys/kiad159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Li G.J., Bressan R.A., Song C.P., Zhu J.K., Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020;62:25–54. doi: 10.1111/jipb.12899. [DOI] [PubMed] [Google Scholar]
- Dai S., Li P., Chen P., Li Q., Pei Y., He S., Sun Y., Wang Y., Kai W., Zhao B., et al. Transcriptional regulation of genes encoding ABA metabolism enzymes during the fruit development and dehydration stress of pear 'Gold Nijisseiki'. Plant Physiol. Biochem. 2014;82:299–308. doi: 10.1016/j.plaphy.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Dang Q., Sha H., Nie J., Wang Y., Yuan Y., Jia D. An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation. Hortic. Res. 2021;8:223. doi: 10.1038/s41438-021-00694-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Chen Y., Liu Z., Liu Z., Shu P., Wang R., Hao Y., Su D., Pirrello J., Liu Y., et al. SlERF.F12 modulates the transition to ripening in tomato fruit by recruiting the co-repressor TOPLESS and histone deacetylases to repress key ripening genes. Plant Cell. 2022;34:1250–1272. doi: 10.1093/plcell/koac025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy I., Sherif S., Mila I., Bouzayen M., Jayasankar S. Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. J. Exp. Bot. 2009;60:907–922. doi: 10.1093/jxb/ern354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciullino A.L., Bidel L.P.R., Urban L. Carotenoid responses to environmental stimuli: integrating redox and carbon controls into a fruit model. Plant Cell Environ. 2014;37:273–289. doi: 10.1111/pce.12153. [DOI] [PubMed] [Google Scholar]
- Gao H. Huazhong Agricultural University; China: 2013. Primary Study on Regulation Mechanism of Carotenoids Accumulation in Citrus Callus. Ph.D. Thesis. [Google Scholar]
- Gao J., Zhang Y., Li Z., Liu M. Role of ethylene response factors (ERFs) in fruit ripening. Food Quality and Safety. 2020;4:15–20. [Google Scholar]
- Gong J., Zeng Y., Meng Q., Guan Y., Li C., Yang H., Zhang Y., Ampomah-Dwamena C., Liu P., Chen C., et al. Red light-induced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22. J. Exp. Bot. 2021;72:6274–6290. doi: 10.1093/jxb/erab283. [DOI] [PubMed] [Google Scholar]
- He Y., Han J., Liu R., Ding Y., Wang J., Sun L., Yang X., Zeng Y., Wen W., Xu J., et al. Integrated transcriptomic and metabolomic analyses of a wax deficient citrus mutant exhibiting jasmonic acid-mediated defense against fungal pathogens. Hortic. Res. 2018;5:43. doi: 10.1038/s41438-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota A., Kato T., Fukaki H., Aida M., Tasaka M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell. 2007;19:2156–2168. doi: 10.1105/tpc.107.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B.Z., Li C.L., Han Y.Y., Shen Y.Y. Characterization of the hot pepper (Capsicum frutescens) fruit ripening regulated by ethylene and ABA. BMC Plant Biol. 2018;18:162. doi: 10.1186/s12870-018-1377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Hu L., Kong W., Yang C., Xi W. Red light-transmittance bagging promotes carotenoid accumulation of grapefruit during ripening. Commun. Biol. 2022;5:303. doi: 10.1038/s42003-022-03270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhao X., Bürger M., Wang Y., Chory J. Two interacting ethylene response factors regulate heat stress response. Plant Cell. 2021;33:338–357. doi: 10.1093/plcell/koaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Li Y., Jia K., Huang B., Dang Q., Wang H., Wang X., Li C., Zhang Y., Nie J., et al. Abscisic acid activates transcription factor module MdABI5-MdMYBS1 during carotenoid-derived apple fruit coloration. Plant Physiol. 2024;195:2053–2072. doi: 10.1093/plphys/kiae188. [DOI] [PubMed] [Google Scholar]
- Jia H., Zuo Q., Sadeghnezhad E., Zheng T., Chen X., Dong T., Fang J. HDAC19 recruits ERF4 to the MYB5a promoter and diminishes anthocyanin accumulation during grape ripening. Plant J. 2023;113:127–144. doi: 10.1111/tpj.16040. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Ji C.Y., Lee C.J., Kim S.E., Park S.C., Kwak S.S. Orange: a target gene for regulating carotenoid homeostasis and increasing plant tolerance to environmental stress in marginal lands. J. Exp. Bot. 2018;69:3393–3400. doi: 10.1093/jxb/ery023. [DOI] [PubMed] [Google Scholar]
- Li B.J., Grierson D., Shi Y., Chen K.S. Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Horticulture Research. 2022;9 doi: 10.1093/hr/uhac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M.H., Li X.Y. Involvement of Transcription Factors and Regulatory Proteins in the Regulation of Carotenoid Accumulation in Plants and Algae. J. Agric. Food Chem. 2023;71:18660–18673. doi: 10.1021/acs.jafc.3c05662. [DOI] [PubMed] [Google Scholar]
- Li Z., Sheerin D.J., von Roepenack-Lahaye E., Stahl M., Hiltbrunner A. The phytochrome interacting proteins ERF55 and ERF58 repress light-induced seed germination in Arabidopsis thaliana. Nat. Commun. 2022;13:1656. doi: 10.1038/s41467-022-29315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Xu Y., Zhang L., Ji Y., Tan D., Yuan H., Wang A. The jasmonate-activated transcription factor MdMYC2 regulates ethylene response factor and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell. 2017;29:1316–1334. doi: 10.1105/tpc.17.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.J., Yin X.R., Xie X.L., Allan A.C., Ge H., Shen S.L., Chen K.S. The Citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein-protein interaction with the vacuolar proton pump, CitVHA-c4. Sci. Rep. 2016;6 doi: 10.1038/srep20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Li L. Carotenoid metabolism: biosynthesis, regulation, and beyond. J. Integr. Plant Biol. 2008;50:778–785. doi: 10.1111/j.1744-7909.2008.00708.x. [DOI] [PubMed] [Google Scholar]
- Ma Q., Ding Y., Chang J., Sun X., Zhang L., Wei Q., Cheng Y., Chen L., Xu J., Deng X. Comprehensive insights on how 2,4-dichlorophenoxyacetic acid retards senescence in post-harvest citrus fruits using transcriptomic and proteomic approaches. J. Exp. Bot. 2014;65:61–74. doi: 10.1093/jxb/ert344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C., Vallarino J.G., Osorio S., Aragüez I., Villarreal N., Ariza M.T., Martínez G.A., Medina-Escobar N., Civello M.P., Fernie A.R., et al. Ethylene is involved in strawberry fruit ripening in an organ-specific manner. J. Exp. Bot. 2013;64:4421–4439. doi: 10.1093/jxb/ert257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.S., Mackinney G., Zscheile F.P. Absorption spectra of alpha and β-carotenes and lycopene. Plant Physiol. 1935;10:375–381. doi: 10.1104/pp.10.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N., Li L., Lu S., Khin N.C., Pogson B.J. Carotenoid metabolism in plants. Mol. Plant. 2015;8:68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Rodrigo M.J., Marcos J.F., Alférez F., Mallent M.D., Zacarías L. Characterization of pinalate, a novel citrus sinensis mutant with a fruit-specific alteration that results in yellow pigmentation and decreased ABA content. J. Exp. Bot. 2003;54:727–738. doi: 10.1093/jxb/erg083. [DOI] [PubMed] [Google Scholar]
- Romero P., Lafuente M.T., Rodrigo M.J. A sweet orange mutant impaired in carotenoid biosynthesis and reduced ABA levels results in altered molecular responses along peel ripening. Sci. Rep. 2019;9:9813. doi: 10.1038/s41598-019-46365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari J.C. Vitamin A and vision. Subcell. Biochem. 2016;81:231–259. doi: 10.1007/978-94-024-0945-1_9. [DOI] [PubMed] [Google Scholar]
- Sandmann G. Diversity and origin of carotenoid biosynthesis: its history of coevolution towards plant photosynthesis. New Phytol. 2021;232:479–493. doi: 10.1111/nph.17655. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Mitsuhara I., Seo S., Ito H., Matsui H., Ohashi Y. Two novel AP2/ERF domain proteins interact with cis-element VWRE for wound-induced expression of the Tobacco tpoxN1 gene. Plant J. 2007;50:1079–1092. doi: 10.1111/j.1365-313X.2007.03111.x. [DOI] [PubMed] [Google Scholar]
- Seale M. Banana ripening control: a non-canonical F-box protein links ethylene and ABA signaling. Plant Physiol. 2022;188:939–940. doi: 10.1093/plphys/kiab560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Liu R., Chen G., Lei J., Ning Z., Tang X., Xiaowan X., Chen M., Cao B., Chen C., et al. Two APETALA2/ETHYLENE RESPONSE FACTORS coordinately with CaMYC2 positively regulate capsaicinoid biosynthesis in pepper (Capsicum annuum) Horticultural Plant Journal. 2023 [Google Scholar]
- Stanley L., Yuan Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front. Plant Sci. 2019;10:1017. doi: 10.3389/fpls.2019.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., He Z., Wei R., Zhang Y., Ye J., Chai L., Xie Z., Guo W., Xu J., Cheng Y., et al. The transcriptional regulatory module CsHB5-CsbZIP44 positively regulates abscisic acid-mediated carotenoid biosynthesis in citrus (Citrus spp.) Plant Biotechnol. J. 2024;22:722–737. doi: 10.1111/pbi.14219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., He Z., Wei R., Yin Y., Ye J., Chai L., Xie Z., Guo W., Xu J., Cheng Y., et al. Transcription factor CsTT8 promotes fruit coloration by positively regulating the methylerythritol 4-phosphate pathway and carotenoid biosynthesis pathway in citrus (Citrus spp.) Hortic. Res. 2023;10 doi: 10.1093/hr/uhad199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Saito D., Hasegawa S., Yamasaki M., Imai M. Vitamin A in health care: suppression of growth and induction of differentiation in cancer cells by vitamin A and its derivatives and their mechanisms of action. Pharmacol. Ther. 2022;230 doi: 10.1016/j.pharmthera.2021.107942. [DOI] [PubMed] [Google Scholar]
- Vranová E., Coman D., Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- Wang W., Fan D., Hao Q., Jia W. Signal transduction in non-climacteric fruit ripening. Hortic. Res. 2022;9 doi: 10.1093/hr/uhac190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ren J., Zhou S., Duan Y., Zhu C., Chen C., Liu Z., Zheng Q., Xiang S., Xie Z., et al. Molecular regulation of oil gland development and biosynthesis of essential oils in Citrus spp. Science. 2024;383:659–666. doi: 10.1126/science.adl2953. [DOI] [PubMed] [Google Scholar]
- Wang G., Wang H., Zhu J., Zhang J., Zhang X., Wang F., Tang Y., Mei B., Xu Z., Song R. An expression analysis of 57 transcription factors derived from ESTs of developing seeds in maize (Zea mays) Plant Cell Rep. 2010;29:545–559. doi: 10.1007/s00299-010-0843-7. [DOI] [PubMed] [Google Scholar]
- Wang X., Zeng W., Ding Y., Wang Y., Niu L., Yao J.L., Pan L., Lu Z., Cui G., Li G., et al. Peach ethylene response factor PpeERF2 represses the expression of ABA biosynthesis and cell wall degradation genes during fruit ripening. Plant Sci. 2019;283:116–126. doi: 10.1016/j.plantsci.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Wu B., Shen F., Wang X., Zheng W.Y., Xiao C., Deng Y., Wang T., Yu Huang Z., Zhou Q., Wang Y., et al. Role of MdERF3 and MdERF118 natural variations in apple flesh firmness/crispness retainability and development of QTL-based genomics-assisted prediction. Plant Biotechnol. J. 2021;19:1022–1037. doi: 10.1111/pbi.13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Xu Z., Zhang Y., Chai L., Yi H., Deng X. An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J. Exp. Bot. 2014;65:1651–1671. doi: 10.1093/jxb/eru044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Ding C., Hu H., Dong G., Zhang G., Qian Q., Ren D. Molecular events of rice AP2/ERF transcription factors. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms231912013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Bao C., Chen P., Cao F., Liu X., Geng D., Li Z., Li X., Hou N., Zhi F., et al. Abscisic acid homeostasis is mediated by feedback regulation of MdMYB88 and MdMYB124. J. Exp. Bot. 2021;72:592–607. doi: 10.1093/jxb/eraa449. [DOI] [PubMed] [Google Scholar]
- Xie Z., Nolan T.M., Jiang H., Yin Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019;10:228. doi: 10.3389/fpls.2019.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.-X., Meng D., Yang Q., Chen T., Qi M., Li X.-Y., Ge H., Chen J.-W. Sorbitol induces flower bud formation via the MADS-box transcription factor EjCAL in loquat. J. Integr. Plant Biol. 2023;65:1241–1261. doi: 10.1111/jipb.13439. [DOI] [PubMed] [Google Scholar]
- Yang Y.-Z., Li T., Teng R.-M., Han M.-H., Zhuang J. Low temperature effects on carotenoids biosynthesis in the leaves of green and albino tea plant (Camellia sinensis (L.) O. Kuntze) Sci. Hortic. 2021;285 [Google Scholar]
- Yin X.R., Xie X.L., Xia X.J., Yu J.Q., Ferguson I.B., Giovannoni J.J., Chen K.S. Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J. 2016;86:403–412. doi: 10.1111/tpj.13178. [DOI] [PubMed] [Google Scholar]
- Yuan H., Zhang J., Nageswaran D., Li L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015;2 doi: 10.1038/hortres.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P., Jiang Z., Sun Q., Wei R., Yin Y., Xie Z., Larkin R.M., Ye J., Chai L., Deng X. Jasmonate activates a CsMPK6-CsMYC2 module that regulates the expression of β-citraurin biosynthetic genes and fruit coloration in orange (Citrus sinensis) Plant Cell. 2023;35:1167–1185. doi: 10.1093/plcell/koac363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jin J., Wang N., Sun Q., Feng D., Zhu S., Wang Z., Li S., Ye J., Chai L., et al. Cytochrome P450 CitCYP97B modulates carotenoid accumulation diversity by hydroxylating β-cryptoxanthin in Citrus. Plant Commun. 2024;5 doi: 10.1016/j.xplc.2024.100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ming R., Khan M., Wang Y., Dahro B., Xiao W., Li C., Liu J.-H. ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechnol. J. 2022;20:183–200. doi: 10.1111/pbi.13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Shi Y., Ma Y., Yang X., Yin X., Zhang Y., Xiao Y., Liu W., Li Y., Li S., et al. The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 2020;18:2267–2279. doi: 10.1111/pbi.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yin X., Xiao Y., Zhang Z., Li S., Liu X., Zhang B., Yang X., Grierson D., Jiang G., et al. An ethylene response factor-MYB transcription complex regulates furaneol biosynthesis by activating quinone oxidoreductase expression in Strawberry. Plant Physiol. 2018;178:189–201. doi: 10.1104/pp.18.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Y., Sun Q., Lu S., Chai L., Ye J., Deng X. Citrus transcription factor CsHB5 regulates abscisic acid biosynthetic genes and promotes senescence. Plant J. 2021;108:151–168. doi: 10.1111/tpj.15431. [DOI] [PubMed] [Google Scholar]
- Zhang D., Zhu K., Shen X., Meng J., Huang X., Tan Y., Cardinale F., Liu J., Li G., Liu J. Two interacting ethylene response factors negatively regulate peach resistance to Lasiodiplodia theobromae. Plant Physiol. 2023;192:3134–3151. doi: 10.1093/plphys/kiad279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Liu X., Gong Q., Cao J., Shen W., Yin X., Grierson D., Zhang B., Xu C., Li X., et al. Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 2021;19:671–688. doi: 10.1111/pbi.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Nie K., Zhou H., Yan X., Zhan Q., Zheng Y., Song C.P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020;228:596–608. doi: 10.1111/nph.16713. [DOI] [PubMed] [Google Scholar]
- Zhao J., Xu Y., Li H., Zhu X., Yin Y., Zhang X., Qin X., Zhou J., Duan L., Liang X., et al. ERF5.1 modulates carotenoid accumulation by interacting with CCD4.1 in Lycium. Hortic. Res. 2023;10 doi: 10.1093/hr/uhad230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Rao S., Wrightstone E., Sun T., Lui A.C.W., Welsch R., Li L. Phytoene synthase: the key rate-limiting enzyme of carotenoid biosynthesis in plants. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.884720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Luo T., Liu C., Wang Y., Yang H., Yang W., Zheng L., Xiao X., Zhang M., Xu R., et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017;216:178–192. doi: 10.1111/nph.14684. [DOI] [PubMed] [Google Scholar]
- Zhu K., Chen H., Mei X., Lu S., Xie H., Liu J., Chai L., Xu Q., Wurtzel E.T., Ye J., et al. Transcription factor CsMADS3 coordinately regulates chlorophyll and carotenoid pools in citrus hesperidium. Plant Physiol. 2023;193:519–536. doi: 10.1093/plphys/kiad300. [DOI] [PubMed] [Google Scholar]
- Zhu K., Sun Q., Chen H., Mei X., Lu S., Ye J., Chai L., Xu Q., Deng X. Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J. Exp. Bot. 2021;72:3137–3154. doi: 10.1093/jxb/erab047. [DOI] [PubMed] [Google Scholar]
- Zou J., Li N., Hu N., Tang N., Cao H., Liu Y., Chen J., Jian W., Gao Y., Yang J., et al. Co-silencing of ABA receptors (SlRCAR) reveals interactions between ABA and ethylene signaling during tomato fruit ripening. Hortic. Res. 2022;9 doi: 10.1093/hr/uhac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.