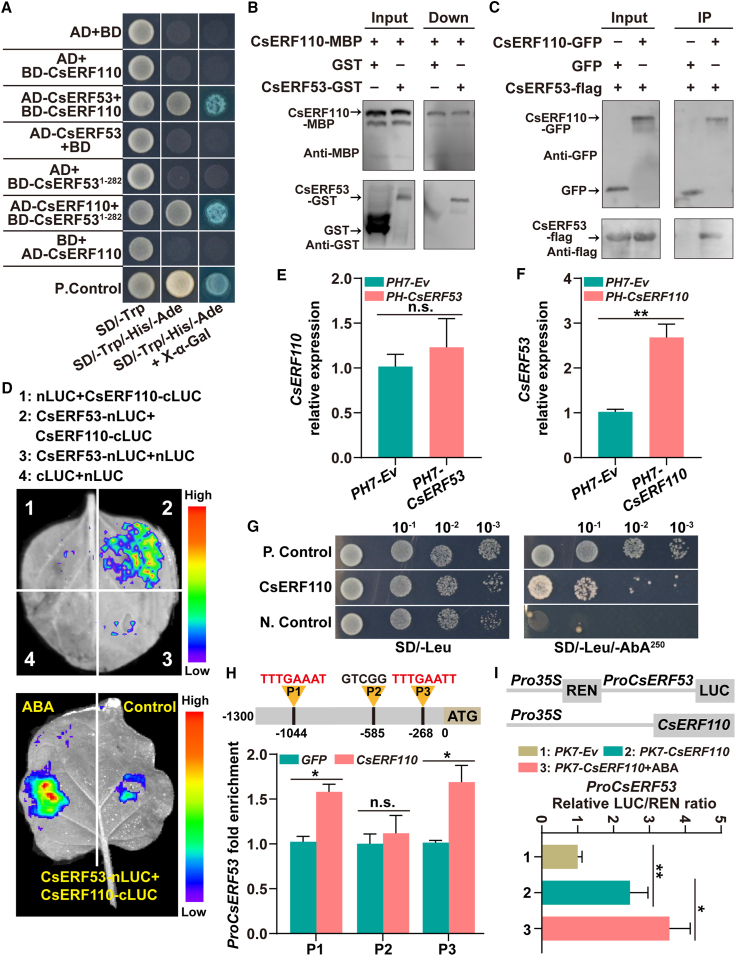
Figure 3.
CsERF110 binds CsERF53 and transcriptionally activates CsERF53.
(A) A Y2H assay reveals an interaction between CsERF53 and CsERF110. Yeast grown in SD/-Trp/-Leu medium and SD/-Trp/-Leu/-His/-Ade medium are shown. The interaction is indicated by yeast growth and X-α-Gal staining.
(B) The interaction between CsERF53 and CsERF110 was analyzed using a pull-down assay. The band detected by the GST antibody in the pull-down protein sample indicates the interaction between CsERF53 and CsERF110.
(C) The interaction between CsERF53 and CsERF110 was confirmed with a coIP assay. GFP antibody beads were used for IP. Anti-GFP and anti-FLAG antibodies were used for immunoblot analyses. The band detected by the GFP antibody in the IP samples indicates an interaction between CsERF53 and CsERF110.
(D) A luciferase complementation imaging assay shows that CsERF53 interacts with CsERF110, and this interaction is enhanced by ABA treatment. Luciferase activity was imaged in the indicated regions 3 days after infiltration. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination.
(E and F) Expression of CsERF110(E) and CsERF53(F) in transgenic calli.
(G) A Y1H assay identified an interaction of CsERF110 with the CsERF53 promoter. PGADT7-Rec-p53+p53-AbAi and empty PGADT7+pAbAi-ProCsERF53 served as the positive (P. Control) and negative (N. Control) controls, respectively. Aureobasidin A was the yeast cell growth inhibitor.
(H) ChIP–PCR assays show the interaction of CsERF110 with several regions in the promoter of CsERF53. The black lines represent the putative binding motifs of ERF family proteins in the CsERF53 promoter. Cross-linked chromatin samples were extracted from GFP-CsERF110 fruit calli and precipitated with an anti-GFP antibody. The eluted DNA fragment was amplified by qPCR.
(I) Dual-luciferase assays indicated that CsERF110 positively regulates CsERF53 transcription, and this effect is enhanced by ABA treatment. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination. Data are presented as means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).