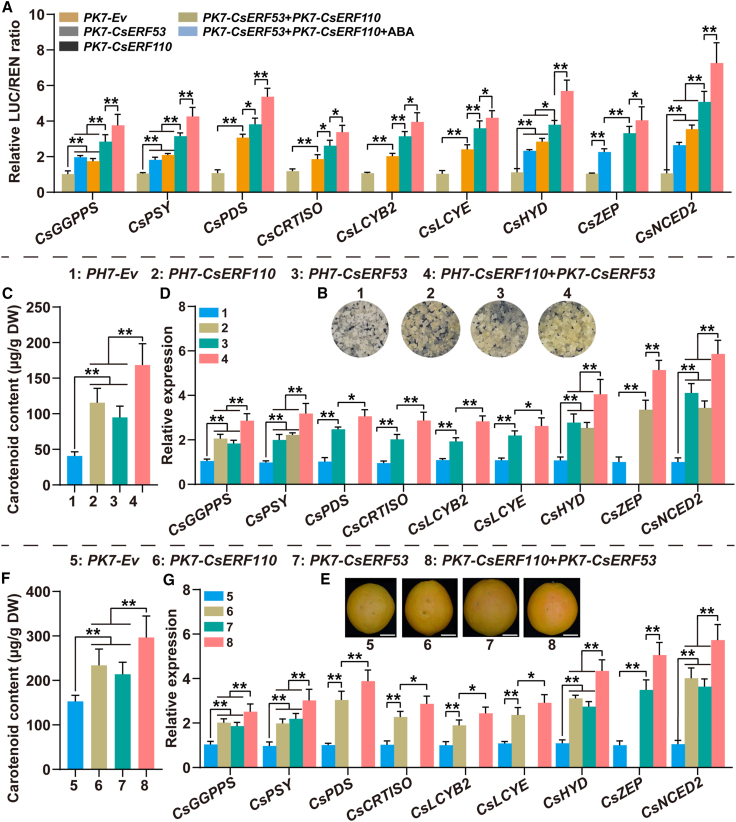
Figure 6.
The transcriptional regulatory module CsERF110–CsERF53 positively regulates carotenoid biosynthesis in citrus.
(A) Effect of the transcriptional regulatory module CsERF110–CsERF53 on the regulation of the promoter activity of downstream target genes. ABA treatment (100 μM) in the dual-luciferase assay was performed for 1 day before determination.
(B–D) Stable transformation of CsERF110 and CsERF53 in citrus calli. PH7-Ev is the control. PH7-CsERF110/CsERF53 indicates overexpression of CsERF110 or CsERF53. PH7-CsERF110+PK7-CsERF53 indicates co-overexpression of CsERF110 and CsERF53. (B) Phenotypes. (C) Carotenoid content (μg/g DW). (D) The expression levels of CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, CsZEP, and CsNCED2.
(E–G) Transient expression of CsERF110 and CsERF53 in citrus fruit. PK7-Ev is the control. PK7-CsERF110/CsERF53 indicates overexpression of CsERF110 or CsERF53. PK7-CsERF110+PK7-CsERF53 indicates co-overexpression of CsERF110 and CsERF53. Scale bars, 3 cm. (E) Phenotypes. (F) Carotenoid content (μg/g DW). (G) Expression levels of CsGGPPS, CsPSY, CsPDS, CsCRTISO, CsLCYB2, CsLCYE, CsHYD, CsZEP, and CsNCED2. Data represent means ± SDs of three biological replicates. Asterisks indicate statistically significant differences determined by Student’s t test (∗p < 0.05; ∗∗p < 0.01; n.s., no significant difference).