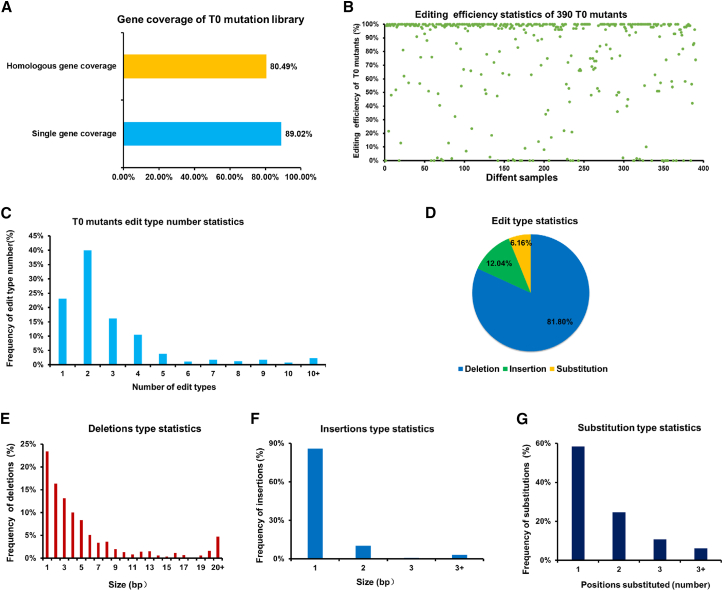
Figure 3.
Molecular detection results for T0-generation plants of the GhCPK mutant library.
(A) Gene coverage of T0-generation plants from the GhCPK mutant library. Yellow represents the coverage of homologous genes, and blue represents the coverage of individual genes.
(B) Editing efficiency of 390 T0-generation plants from the GhCPK mutant library. Different samples are represented by green dots.
(C) Types of edits in T0-generation plants of the GhCPK mutant library. “Edit type” refers to different types of mutated reads produced after genes have been edited by the Cas9 protein. The number of edit types varies for each plant, ranging from one to more than 10.
(D) Classification of edit types in T0-generation plants of the GhCPK mutant library. Blue represents base absence, green represents base insertion, and yellow represents base substitution.
(E) Base deletion statistics. The x axis represents the number of deleted bases, and the y axis represents the frequency.
(F) Base insertion statistics. The x axis represents the number of inserted bases, and the y axis represents the frequency.
(G) Base substitution statistics. The x axis represents the number of substituted bases, and the y axis represents the frequency.