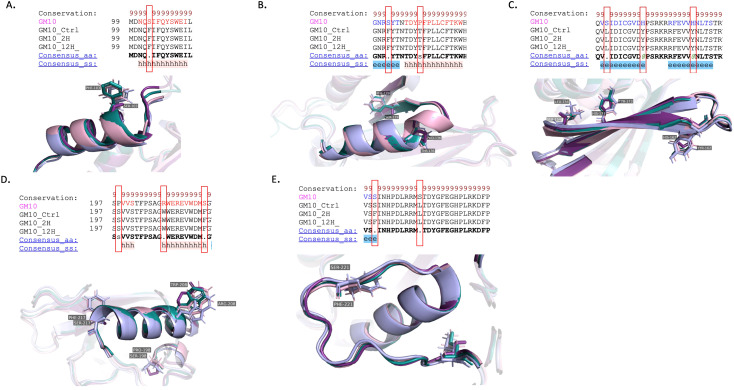
Figure 10.
Structural analysis of RNA editing variants in the NAD9 protein of the Triticum aestivum Gemmiza10 cultivar across the three drought treatment time points (control, 2 hours, 12 hours). (A–E) The panels display a structural alignment of the NAD9 protein virtual translated from DNA (GM-DNA) in comparison with highlighting RNA editing-induced changes in samples collected from the control (GM-C), 2-hour (GM-2H) and 12-hour (GM-12H) post drought treatments. (A) Structural comparison of an alpha-helix region across the three time points, illustrating the substitution of an amino acid from the control sequence and its conservation across the treated samples. (B) Visualization of another alpha-helical segment where RNA editing modifies residues. (C) Beta-strands featuring multiple editing sites, indicating extensive structural alterations post-RNA editing. (D) A conserved alpha-helix with RNA editing sites. (E) C-terminal beta-strand region, with mapped RNA editing sites.