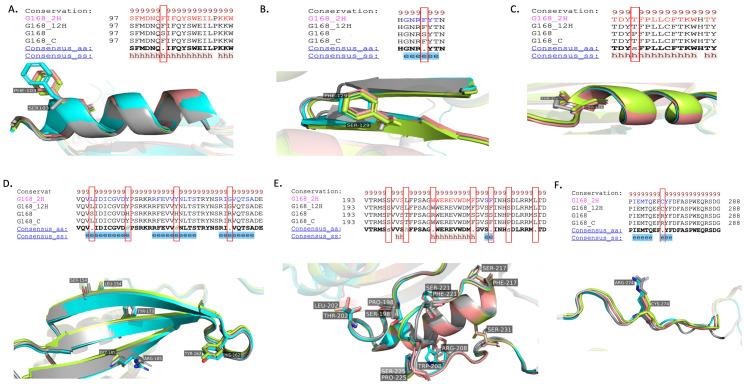
Figure 9.
Structural analysis of RNA editing variants in the NAD9 protein of the Triticum aestivum Giza168 cultivar across the three drought treatment time points (control, 2 hours, 12 hours). (A-F) Structural variants resulting from RNA editing in the NAD9 protein of the Triticum aestivum Giza168 (G168) cultivar were analyzed through alignment with the NAD9 protein virtual translated from DNA (G168_ gray) and three additional models: NAD9_G168_control (G168_C) in cyan, NAD9_G168_2H (G168_2H) in the Limon, and NAD9_G168_12H (G168_12H) in the dark salmon. (A) Detailed view of an alpha-helix region showing conserved RNA editing sites across different treatment time points (control, 2 hours, 12 hours). (B) Ribbon diagrams of the protein structure around RNA editing sites with accentuated changed residues, illustrate the potential impact on the local structure. (C) no-effect in alpha helix integrity due to RNA editing. (D) Beta-strand representation of RNA editing sites showing the conservation of structural changes. (E, F) C-terminal region alignment, emphasizing the conserved RNA editing sites and their structural significance.