Figure 4.
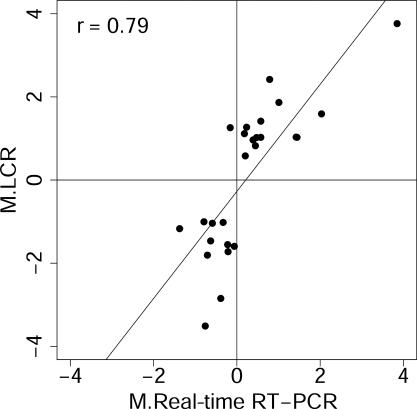
LCR targets detect differential expression that is missed by oligo(dT)-primed targets. Differential gene expression detected by LCRs but not by oligo(dT) targets are confirmed by quantitative RT–PCR. This result indicates that LCRs can be used to detect changes in gene expression that cannot be detected using an oligo(dT) target on these microarrays. Limma output for these genes is in Supplementary Table 3, while accession numbers, current Unigene designations, and descriptions are in Supplementary Table 4. Axis label ‘M.LCRs’ is the average log2 ratio of the normalized channel intensities, I, for the 1 and 0 h treatments reported by the LCR targets, i.e. M.LCRs = log2(It=1/It=0), and ‘M.Real-time RT–PCR’ is the log2 of the corresponding ratio reported by real-time RT–PCR. Pearson's correlation, r, is shown.