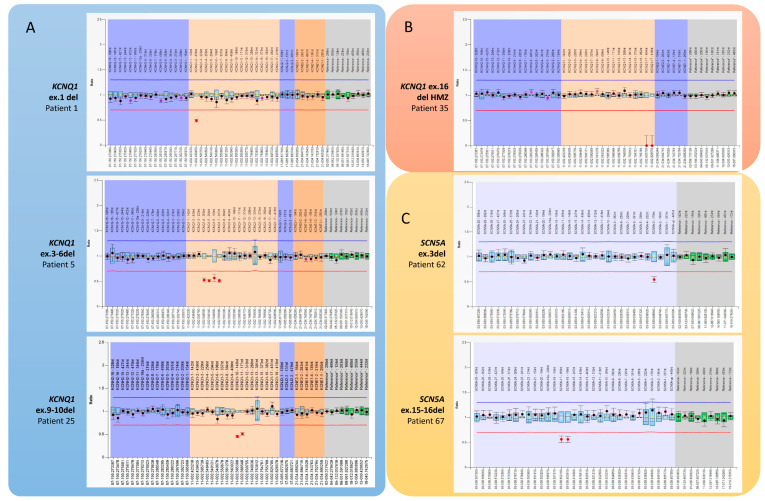
Figure 4.
Detection of CNVs involving arrhythmic genes identified by P114 and P108 MRC Holland MLPA kits. The relative peak ratio of MLPA products in these profiles reflects the presence of exon deletions or duplications compared with reference samples. Blue/green bars represent a 95% confidence interval over the reference samples (N = 4), and dots with lines represent a 95% confidence interval estimate for each probe. The chromosomal positions and bands in the MLPA kits are all based on the hg18 genome; we convert the genomic coordinates to hg19 according to the CNVs described in the literature. For this reason, The NM_ sequence used by the company to determine a probe’s ligation site does not always correspond to the exon numbering obtained from the LRG sequences (Locus Reference Genomic). (A,B) We identified different KCNQ1 deletions in 4 unrelated families. (A) Patient 1 shows a single copy loss of exon 1 in KCNQ1 transcript variant 2 (NM_181798.1). (B) In patient 35, we identified a homozygous deletion of KCNQ1 exon 16. (C) Deletion in SCN5A gene.