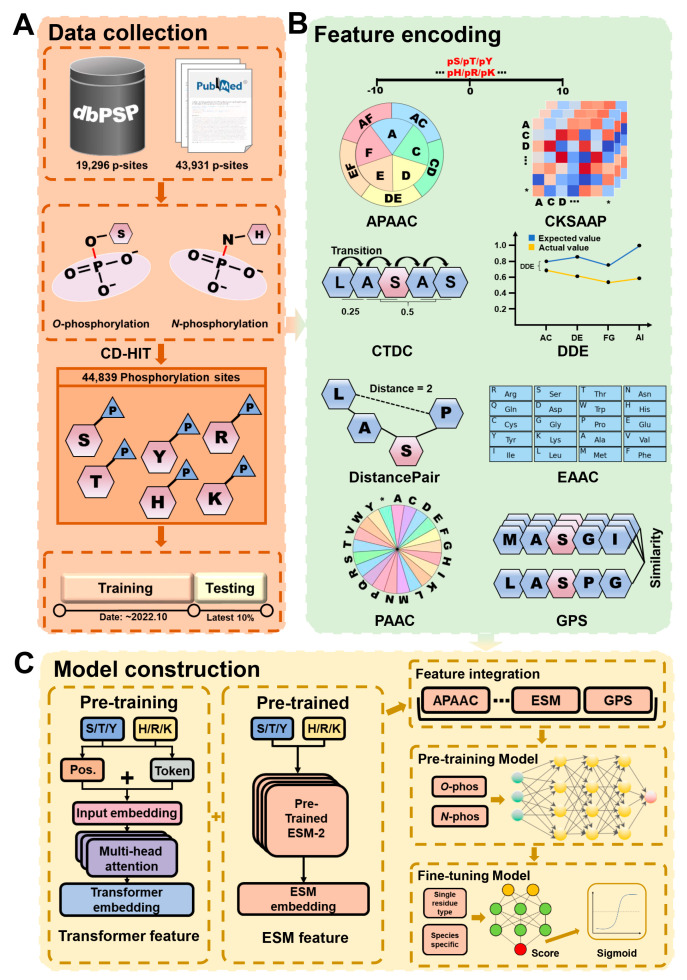
Figure 1.
The procedure for the development of the GPS-pPLM, including data collection, feature encoding, and model construction. (A) Data preparation of p-sites curated from the literature and dbPSP 2.0. (B) Sequence feature encoding of the GPS-pPLM algorithm. In total, 8 types of sequence features were used, including APAAC, CKSAAP, CTDC, DDE, distance pair, EAAC, PAAC, and GPS. (C) Model construction methods used for GPS-pPLM. Two machine learning approaches, including transformer and DNN, were used for the construction of predictive models. We pretrained two general models, the O-phosphorylation model and the N-phosphorylation model, and then the pretrained models were fine-tuned using data specific to a single-residue type or species.