Figure 3.
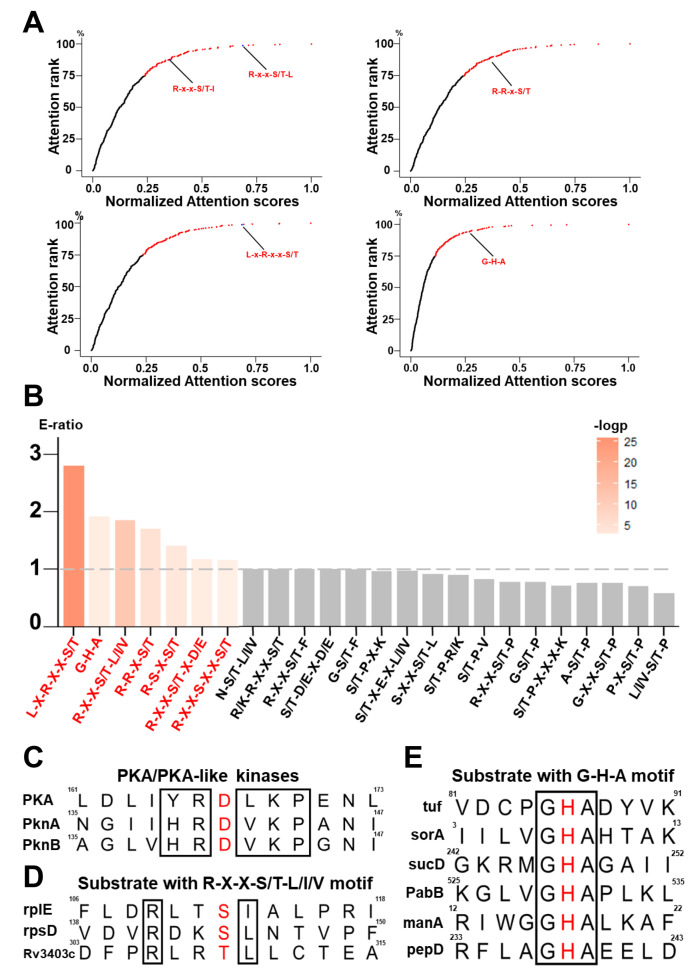
Identification of prokaryotic p-site motifs. (A) Eukaryotic-like phosphorylation motifs identified in prokaryotes through the attention mechanism, including R-X-X-S/T-L/I/V, R-R-X-S/T, L-X-R-X-X-S/T, and G-H-A. (B) The enrichment results of 24 motifs containing two or more amino acids were obtained from all training data (p-value < 0.01). These motifs were identified via the motif-x tool from the phosphoproteomic data of 48 eukaryotes. (C) Sequence alignment results of the PKA, PknA, and PknB kinase functional sites. (D) The substrates that can be specifically phosphorylated by PknB include R-X-X-S/T-L/I/V motifs in prokaryotes. (E) Display of sequences near the p-site of the G-H-A motif in the training dataset.