Table 1.
Chemical structures, design strategies, and functions of ABA signaling modulators.
| Category | Designation | Chemical Structure | Design Strategy | Function | References |
|---|---|---|---|---|---|
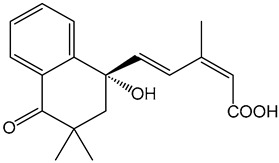
| ABA analogs | 2′,3′-iso-PhABA |
|
The introduction of a benzo ring at the 2′, 3′ position alters the molecular conformation. | Inhibitory effects on lettuce (Lactuca sativa) and Arabidopsis seed germination, wheat germination, and rice seedling elongation. | [175] |
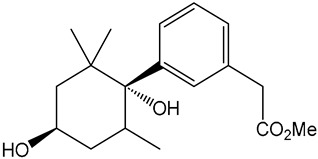
| (+)-BP2A |
|
The diene on the side chain of ABA is replaced by a phenylacetic acid group. | ABA-like activity that inhibits the germination of tomato, lettuce, and rice seeds. | [177] | |
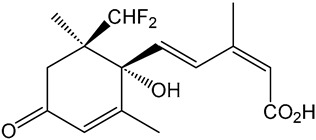
| 9′,9′-difluoro-ABA |
|
Chemical modifications to stabilize the cyclohexanone ring. | In radish seedlings and rice seedlings, the deactivation rate of ABA analogs is slower than that of ABA. | [180] | |
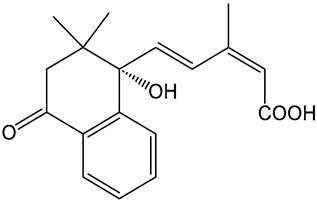
| (+)-tetralone ABA |
|
Fusion of a cyclohexenone ring with a benzene ring. | More effectively complements the growth retardation of ABA-deficient Arabidopsis mutants than natural ABA. | [178] | |
| ABA agonists | Quinabactin |
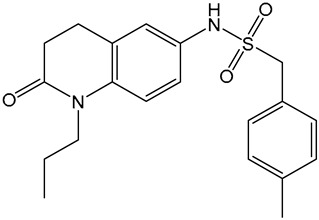
|
Hydrogen bonds or halogen bonds form hydrophobic interactions with amino acid residues of the ABA receptor. | An inhibitory effect on seed germination. | [181] |
| Opabactin |
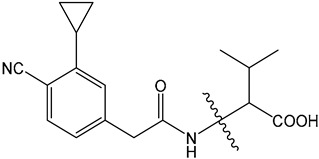
|
Structure-based design and molecular docking screening. | Inhibits seed germination and seedling growth in Arabidopsis and rice. | [182] | |
| JFA |
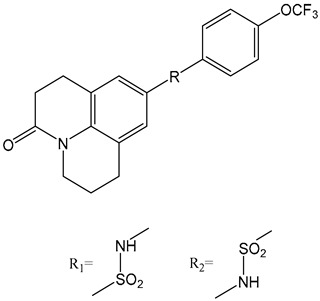
|
Cell-free drug screening system for wheat. | Activation of PYR1 and PYL1 inhibits seed germination and cotyledon greening in seedlings. | [183] | |
| ABA antagonists | (+)-PAT |
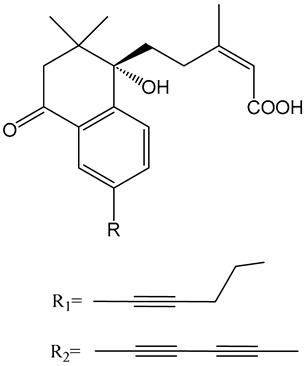
|
Conformational constraint method. | Inhibits ABA-induced seed germination in Arabidopsis. | [184] |
| AAT1 |
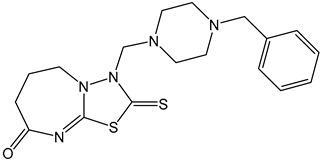
|
Chemical Genetics Screening | Broad-spectrum Arabidopsis ABA receptor antagonist. | [185] | |
| Aantabatin |
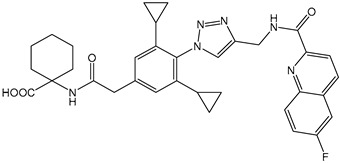
|
Click chemistry for creating a derivative library. | Accelerates seed germination in Arabidopsis, tomato, and barley. | [186] |
