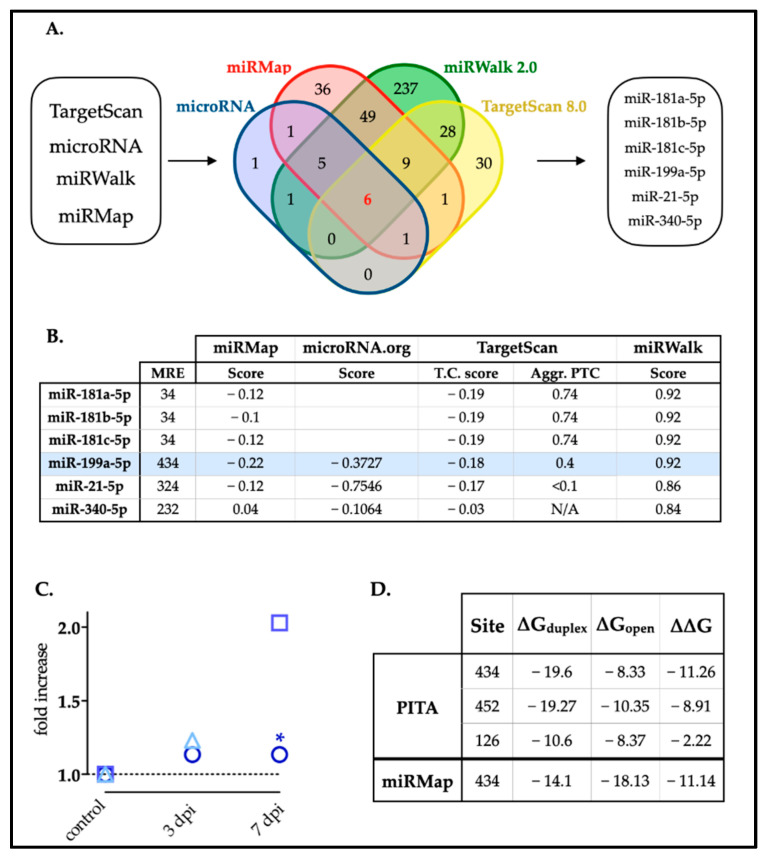
Figure 1.
Selection of miRNAs with predicted MREs in the rat 3′-UTR-XIAP. (A) Venn diagram representing the number of miRNAs predicted by each of the four algorithms (TargetScan 8.0, miRanda, miRWalk and miRMap); or by more than one. Six miRNAs candidates were predicted by all four algorithms: miR-181a-5p, miR-181b-5p, miR-181c-5p, miR-199a-5p, miR-21-5p, and miR340-5p. (B) The table shows the selected miRNAs and the prediction scores calculated by each algorithm. miR-199a-5p is highlighted in blue. (C) Summary graph illustrating miR-199a-5p fold increase data at 0, 3, and 7 dpi (days post-injury) from our previous study and others. Circles represent data from Yunta et al. [6], * p < 0.05 at 7 dpi; squares represent data from Liu et al. [33]; and triangles represent data from Chen et al. [32]. (D) Localization of the different MREs for miR-199a-5p in the whole sequence of the rat 3′UTR-XIAP, indicating the ∆∆G score for miRNA–target interactions, computed as the free energy gained by transitioning from the state in which the miRNA and the target are unbound (∆G open) and the state in which the miRNA binds its target (∆G duplex), according to PITA algorithm.