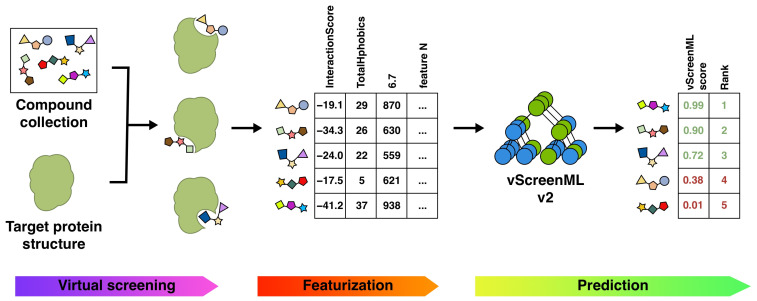
Figure 4.
Incorporating vScreenML 2.0 into a virtual screening pipeline. Candidate protein–ligand complexes can be generated by receptor-based methods (i.e., docking or pharmacophoric alignment to one or more known ligands). Scoring each of the resulting complexes with vScreenML 2.0 involves extracting “features” from each model using the PyRosetta (v. 2023.14+release.7132bdc), Binana (v.2.1), RF-Score (v1), RDKit (v.2023.9.5), LUNA (v.0.13.0), and PocketDruggability (v.0.98.1) packages. These features are used as input to vScreenML 2.0, which uses an XGBoost-based model to produce a single score reflecting the prioritization of the compounds in the screening collection.