Abstract
Red beetroots, rich in betanin, may act as prebiotics and impact gut microbiota. Because the human gut microbiota is unique to each person, the effectiveness of prebiotics varies with the enterotype. In this study, we hypothesized that the effects of red beetroot powder (RP) and betanin pigment (BP) would differ depending on the enterotype. Fecal samples from 30 subjects were analyzed and categorized into three enterotypes: Phocaeicola, Prevotella, and Bifidobacterium. Feces were collected from one representative subject from each enterotype cluster for fermentation. Results showed that RP and BP affected microbiota composition and short-chain fatty acid (SCFA) production differently across enterotypes. The Bifidobacterium cluster showed significantly reduced alpha diversity, with the direction of change in the gut microbiota composition being different from that of other subjects. Additionally, SCFAs significantly increased, with the highest increase in the Bifidobacterium cluster. In this cluster, metabolic pathways related to SCFAs (i.e., starch and sucrose metabolism and glycolysis/gluconeogenesis) were altered. Conversely, Prevotella-dominant feces exhibited fewer changes in SCFAs and a lower increase in Bifidobacterium abundance than the others. These findings highlight that RP and BP elicit enterotype-specific responses in the gut microbiota composition and SCFA production, emphasizing the importance of enterotypes in personalized nutrition.
Keywords: Beta vulgaris L., gut microbiota, enterotype, short-chain fatty acid, personalized nutrition
1. Introduction
Trillions of microorganisms reside in the human intestine, and the intestinal microbiota is often referred to as the second genome [1]. The gut microbiota interacts with the host, contributing significantly to overall health and playing a crucial role in digestion and nutrient absorption. For example, the gut microbiota is involved in metabolic diseases such as obesity, hypertension, diabetes, atopy, and depression [2,3,4,5]. These microbes affect the immune system, break down food to produce nutrients, and metabolize food to produce various metabolites such as essential amino acids, short-chain fatty acids (SCFAs), and vitamins [6]. SCFAs are metabolites produced by the gut microbiota during the fermentation of dietary fiber and non-digestible carbohydrates, boosting the immune system, thereby increasing resistance to infection and inflammation, and affecting the nervous and endocrine systems by acting as signaling molecules [7,8]. Importantly, the composition of the gut microbiota is closely correlated with SCFA production [9]. Specific bacterial groups, such as Faecalibacterium, Bifidobacterium, and Bacteroides, are particularly effective at fermenting dietary fibers, resulting in higher levels of SCFAs. Conversely, dysbiosis—an imbalance in gut microbiota—can lead to decreased SCFA production.
The human gut microbiota is strongly influenced by a variety of factors, including age, nutritional status, and geographic environment. Notably, enterotypes are defined as various clusters of the human gut microbiota, categorized based on core bacteria such as Bacteroides, Prevotella, Ruminococcus, Bifidobacterium, and Faecalibacterium [10,11,12]. Interestingly, recent research suggests that individuals exhibit different metabolic responses, even when consuming the same diet, depending on their enterotypes [13]. Accordingly, enterotypes are important indicators of personalized nutrition. Personalized nutrition is a nutritional strategy that considers each individual’s genetics, health status, lifestyle, and dietary habits to customize their nutrient requirements and intake recommendations. Recently, microbiome analysis methods, such as enterotyping, have been introduced to minimize individual differences and identify the effects of diet [14]. Thus, predicting dietary responses based on enterotypes can help develop personalized nutritional strategies that consider an individual’s microbiome structure, which is expected to play an important role in personalized medicine and nutrition.
Red beetroot (Beta vulgaris L.) is a vegetable rich in phytochemicals such as dietary fiber, polyphenols, and betalains, making it popular worldwide for its health benefits. The main betalain in red beetroot is betanin, which belongs to the betacyanin subgroup and is the only betacyanin approved as a food additive by the European Union and the Food and Drug Administration (FDA) [15,16]. Betanin possesses anti-inflammatory, antioxidant, and immune-regulating effects [17]. Remarkably, these beneficial effects may be mediated by gut microbiota [18]. Enzymes produced by gut microbiota, such as β-glucosidase and glycoside hydrolases, can metabolize phytochemicals, including dietary fiber, polyphenols, and betanin, into SCFAs [19,20,21]. A previous study reported that the consumption of red beetroot juice increases SCFAs, potentially associated with an increase in betacyanin catabolites by gut microbiota [22]. Additionally, consuming whole beetroot has been shown to lower systolic blood pressure and enhance SCFA production through the regulation of intestinal microbiota in elderly individuals [23]. Thus, red beetroot and betanin can be utilized as prebiotics. However, it is important to note that representative bacteria, such as Bifidobacterium and Bacteroides, which produce microbial enzymes capable of metabolizing these substances in the gut, are core bacteria of specific enterotypes [24,25]. Therefore, the response to red beetroot and betanin may vary depending on the enterotype.
This study aimed to provide basic information for the development of personalized prebiotic materials by investigating the effects of red beet powder (RP) and betanin pigment (BP) on the gut microbiota composition and short-chain fatty acid production based on enterotype using a gastrointestinal digestion and fecal fermentation (GID-FF) model. Our findings highlight that RP and BP elicit enterotype-specific responses in the gut microbiota composition and SCFA production, emphasizing the importance of enterotypes in personalized nutrition.
2. Materials and Methods
2.1. Reagents and Materials
RP was prepared using beetroot grown in the Jeju Special Self-Governing Province Agricultural Research and Extension Service. BP, α-amylase, pepsin, pancreatin, and standard SCFAs (acetate, propionate, and butyrate) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.2. Test Subjects and Fecal Sample Collection
This study was approved by the Institutional Review Board of Jeju National University (approval number: JJNU-IRB-2024-026) and registered with the Clinical Research Information Service (CRIS: https://cris.nih.go.kr/cris/index/index.do, Clinical Trial Registry Number: KCT0009657, accessed on 26 July 2024). We recruited 30 healthy subjects (17 men and 13 women) for enterotype analysis (primary outcome) with no underlying medical conditions such as diabetes, cancer, liver disease, or history of gastrointestinal disease. Based on this, we selected one subject per enterotype to assess the impact on gut microbiota (secondary outcomes). None of the individuals had been treated with antibiotics for at least 3 months before sample collection. Fecal samples were stored at −20 °C immediately upon collection and subsequently transported to the lab for storage at −80 °C. Fecal samples were collected immediately for fermentation.
2.3. In Vitro Gastrointestinal Digestion (GID) and Fecal Fermentation (FF)
GID-FF was performed as previously described, with several modifications [26,27]. In the salivary digestion phase, 0.5 g of RP and BP were mixed with 4.5 mL of PBS (pH 7), 3.5 mL of simulated salivary fluid (SSF), 0.5 mL of alpha-amylase (1500 U/mL make-up SSF), 25 μL of 0.3 M CaCl2, and 975 μL of distilled water (DW). This was incubated at 37 °C for 2 min at 150 rpm. In the gastric digestion phase, the salivary digestion product was mixed with 7.5 mL of simulated gastric fluid (SGF), 1.6 mL of pepsin (25,000 U/mL in SGF), 5 μL of 0.3 M CaCl2, and 695 μL of DW, adjusted to pH 2 using 4 N HCl, and incubated at 37 °C for 2 h at 100 rpm. In the intestinal digestion phase, the gastric digestion product was mixed with 11 mL of simulated intestinal fluid (SIF), 5 mL of pancreatin (800 U/mL in SIF), 2.5 mL of bile salt, 40 μL of CaCl2, and 1.31 mL of DW, adjusted to pH 7 using 1 M NaOH and incubated at 37 °C for 2 h at 100 rpm.
The day before the fecal fermentation experiment, PBS and the basal culture medium were placed overnight in an anaerobic chamber (90% N2, 5% H2, and 5% CO2) to remove oxygen. The fecal samples were collected in a sterilized 50-mL conical tube and immediately transferred into an anaerobic chamber upon receipt. The transferred fecal sample was homogenized with PBS (20% w/v) using a vortex and sieved through pore sizes of 250, 150, and 25 μm to filter the residue. For fecal fermentation, 1.2 mL of basal culturing medium and 150 μL of GID product (10%) were dispensed into each well of a 96-well deep plate and inoculated with 150 μL of sieved feces in PBS (10%). The fermentation was performed in triplicates at 100 rpm for 6 h at 37 °C in an anaerobic chamber using digital shakers.
2.4. Analysis of Microbial Community
An amount of 1.5 mL of the sample was centrifuged at 13,000 rpm for 5 min at 4 °C. The pellet was used for genomic DNA extraction using a QIAamp PowerFecal Pro DNA Kit (QIAamp, Germantown, MD, USA). To analyze the microbial community in the samples, polymerase chain reaction (PCR) was used to amplify the V3–V4 hypervariable region of the 16S rRNA gene. The PCR product was purified, and individual indices were added to the amplicons for each sample using PCR once again. After purifying the PCR products in the same manner, sequencing was performed using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). All sequencing procedures were performed by Macrogen, Inc. (Seoul, Republic of Korea).
Sequencing data were analyzed according to the MOTHUR SOP guidelines using the MOTHUR software version 1.47.0 (https://mothur.org/wiki/miseq_sop/, accessed on 26 July 2024) [28]. Briefly, raw reads obtained from Miseq were assembled using “make.contigs” and aligned to the SILVA database version 138 using “align.seqs” [29]. Rare sequences and singletons were removed using “pre.cluster” and “spit.abun,” potential chimeric sequences were read using “chimera vserach” [30], taxonomy classification of bacteria was read with Ribosome database project version 18 using “classify.seqs” [31], and undesired taxa sequences (i.e., chloroplast, mitochondria, unknown, and Eukaryota) were eliminated using “remove.lineage” and clustered based on OptiClust algorithms with 97% similarity using “opti.clust” [32]. The number of reads was normalized to 20,000 for downstream analysis. Enterotype classification was performed using “get.communitytype” using the k-means clustering algorithm with tree analysis.
The α-diversity indices for richness and evenness were calculated based on Chao and Shannon within MOTHUR. The β-diversity indices for non-metric multidimensional scaling (NMDS) analysis evaluated the differences in each group. The metabolic pathways of fecal microbiota were estimated using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) version 2.4.2 [33], and KO abundances were converted to KEGG pathway abundances using the ggpicrust2 package version 1.7.3 in R [34].
2.5. SCFAs Extraction and Quantification
An amount of 1.5 mL of the sample was centrifuged at 13,000 rpm for 5 min at 4 °C, and the supernatant was used for SCFA extraction. An amount of 200 μL of the supernatant was added to 800 μL of absolute methanol and homogenized for 2 min. The pH of the mixture was adjusted to 2–3 using HCl, and the mixture was incubated at room temperature for 10 min, with frequent homogenization every 3 min. Finally, the mixture was filtered through a membrane with a pore size of 0.45 μm.
Quantification of SCFAs (acetate, propionate, and butyrate) was conducted using Gas Chromatography (GC2010, Shimadzu, Japan) with a flame ionization detector (FID) on Split mode (10:1 ratio) using a DB-FFAP column (30 m × 0.25 μm × 0.25 μm, Agilent, Santa Clara, CA, USA). The temperature of the inlet and FID was maintained at 230 °C and 280 °C, respectively, and 1 μL of the filtered mixture was injected into the column. During operation, the column oven had the following temperature: 80 °C for 3 min, and then gradually increased to 200 °C at a rate of 15 °C/min. At 200 °C, it was held for another 3 min, then increased to 230 °C at a rate of 5 °C/min, and finally held for 10 min.
2.6. Statistical Analysis
Data is expressed along with standard deviation. SCFAs and α-diversity indices were evaluated using analysis of variance (ANOVA). Analyses of molecular variance (AMOVA), NMDS, and enterotype clustering were performed using the Bray–Curtis coefficient [35]. The differential abundance of microorganisms between groups was calculated using a linear discriminant analysis effect size (LEfSe) based on the Kruskal–Wallis (KW) sum-rank test [36]. Additionally, the significant differential abundance of the predicted microbial metabolic activities between groups was investigated using a two-sided Welch’s t-test via STAMP version 2.1.3 [37]. Correlation analysis between the bacterial genera and SCFAs was performed using Pearson’s rank correlation coefficient.
3. Results
3.1. Enterotype Analysis
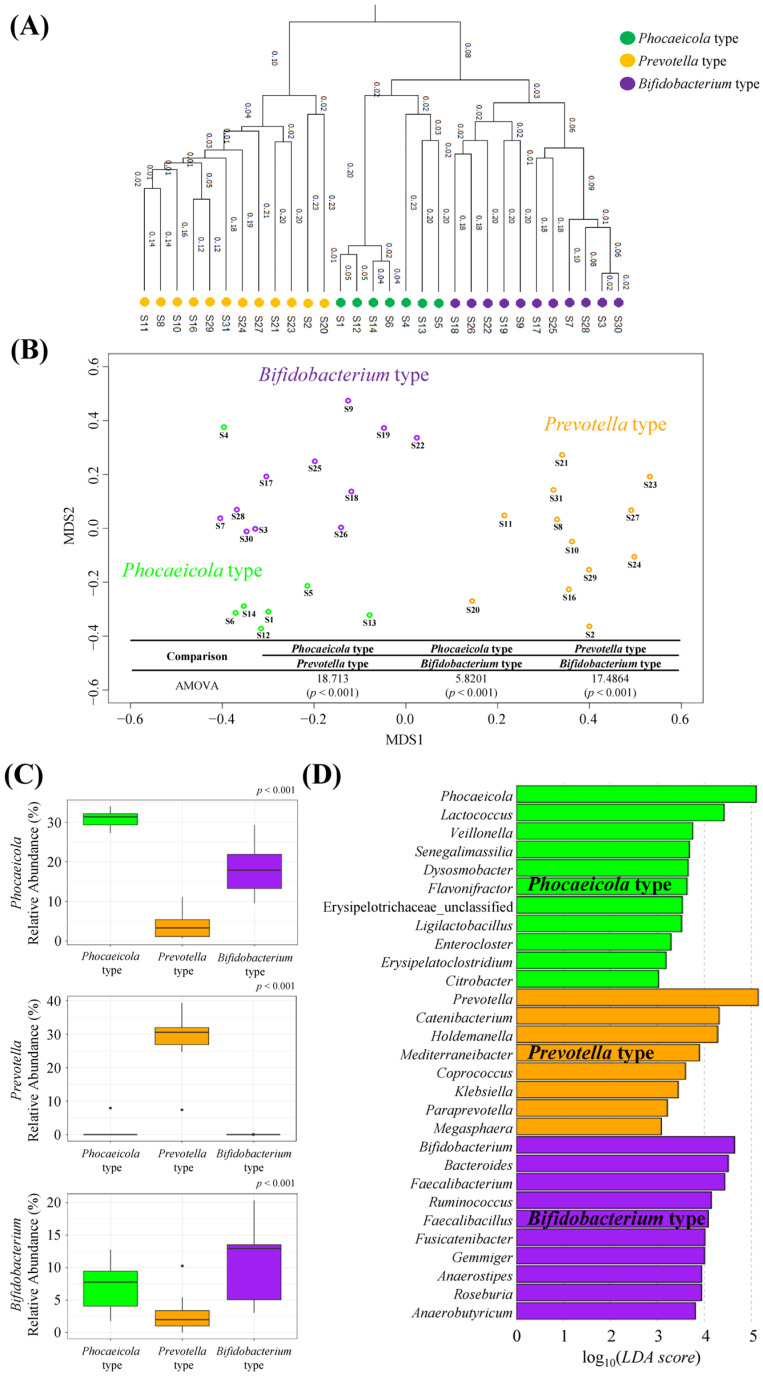
Enterotype analysis was performed on fecal samples collected from 30 participants (participants 1–30). The gut microbiota of the 30 subjects was categorized into three types. Each enterotype not only exhibited a relatively scattered distribution in the Tree and NMDS plots but also demonstrated statistically significant differences, primarily driven by the genera Phocaeicola, Prevotella, and Bifidobacterium as the core microbiota (p < 0.05, Figure 1A–C). We designated enterotypes according to the core microbiota as the Phocaeicola (n = 7), Prevotella (n = 12), and Bifidobacterium (n = 11) types. Analysis of 30 fecal samples at the phylum level revealed that the Phocaeicola type had a higher relative abundance of Proteobacteria than other types. Compared to the other types, the Prevotella type showed a higher relative abundance of Bacteroidetes, whereas the Bifidobacterium type had a higher relative abundance of Firmicutes and Actinobacteria (Figure S1). Furthermore, we examined the microbiota with significantly different genera by enterotype, in addition to the core microbiota, which is summarized in Figure 1D. The Phocaeicola type exhibited enrichment of 11 genera, such as Lactococcus and Veillonella, whereas the Prevotella type demonstrated enrichment of 8 genera, including Catenibacterium and Holdemanella. In contrast, the Bifidobacterium cluster was enriched with 10 genera, including Bacteroides, Ruminococcus, and Faecalibacterium. Characteristics such as age and BMI of the subjects in each type were not significantly different (p > 0.05, Table S1).
Figure 1.
Fecal microbial enterotype clustering. (A) Tree analysis, (B) non-metric multidimensional scaling (NMDS) with analysis of molecular variance (AMOVA), (C) relative abundance of Phocaeicola, Prevotella, and Bifidobacterium in each enterotype, and (D) linear discriminant analysis (LDA) effect size (LEfSe) of three different enterotypes at the genus level. (p < 0.05, LDA score > 3).
3.2. Microbial Communities of GID-FF-Treated Fecal Samples
For GID-FF, fecal samples were collected from one subject of each type from the 30 subjects (Subject 1, Subject 2, and Subject 3 were selected), and the preferentially selected samples (labeled S1-Phocaeicola, S2-Prevotella, and S3-Bifidobacterium) were checked to ensure that they were representative of each type. As shown in Figure S2, the S1-Phocaeicola sample belonged to the Phocaeicola type, whereas S2-Prevotella and S3-Bifidobacterium belonged to the Prevotella and Bifidobacterium types, respectively. In addition, the abundance of Phocaeicola, Prevotella, and Bifidobacterium, which are the core bacteria in each selected subject, was consistent (Figure 2A). Therefore, the selected sample remained representative of each enterotype.
Figure 2.
The fecal microbiota of the subjects selected based on enterotype before fecal fermentation. (A) relative abundance of Phocaeicola, Prevotella, and Bifidobacterium in each subject, and (B) linear discriminant analysis (LDA) effect size (LEfSe) at the genus level (p < 0.05, LDA score > 3).
At the genus level, 17, 18, and 15 genera were enriched in S1-Phocaeicola, S2-Prevotella, and S3-Bifidobacterium samples, respectively (Figure 2B). In particular, Lactococcus, Senegalimassilia, Ligilactobacillus, and Veillonella, which are abundant in the Phocaeicloa type, were enriched in the S1-Phocaeicloa sample, whereas Coprococcus and Mediterraneibacter, which are abundant in the Prevotella type, were enriched in the S2-Prevotella sample. Similarly, Faecalibacterium, Faecalibacillus, Fusicatenibacter, Ruminococcus, Roseburia, and Anaerobutyricum, which are abundant in the Bifidobacterium enterotype, were enriched in the S3-Bifidobacterium sample. At the phylum level, Proteobacteria, which had a higher relative abundance in the Phocaeicloa type, and Actinobacteria, which had a higher relative abundance in the Bifidobacterium type, were both enriched in S1-Phocaeicloa, and Verrucomicrobia were enriched in S2-Prevotella (Figure S3).
3.3. Enterotype-Specific Effects of RP and BP on Gut Microbiota
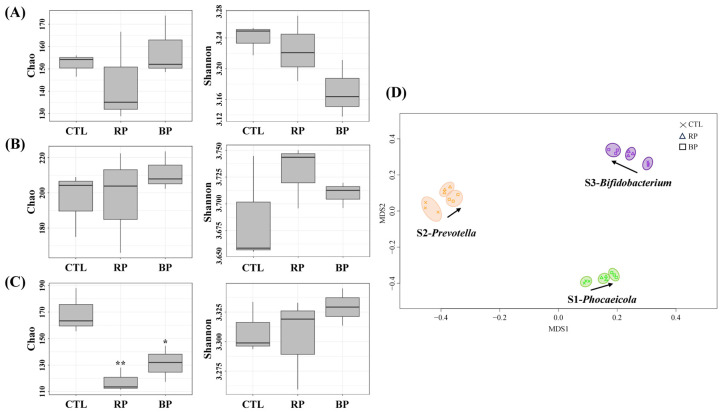
To investigate the effects of RP and BP on the gut microbiota based on enterotype, GID-FF was performed on S1-Phocaeicola, S2-Prevotella, and S3-Bifidobacterium fecal samples treated with RP and BP. The alpha and beta diversities of the fecal samples after fermentation are shown in Figure 3. We compared the Chao and Shannon indices, which represent species richness and evenness of the gut microbiota, respectively. When the S1-Phocaeicola type sample was treated with RP and BP, the Chao indices were 143.52 ± 11.69 and 158.19 ± 7.90 and the Shannon indices were 3.18 ± 0.02 and 3.16 ± 0.03, respectively, while for the S2-Prevotella type sample, the Chao indices were 197.44 ± 16.61 and 211.28 ± 6.36 and the Shannon indices were 3.74 ± 0.05 and 3.71 ± 0.01, respectively. For the S3-Bifidobacterium sample, the Chao indices were 117.76 ± 5.21 and 131.33 ± 7.83, and the Shannon indices were 3.30 ± 0.01 and 3.32 ± 0.02, respectively. Overall, the Chao index was significantly reduced by the RP and BP only in the S3-Bifidobacterium sample (p < 0.05), whereas the Shannon index showed no significant changes in any of the samples (p > 0.05). In the NMDS, no significant changes in the gut microbiota were observed after fecal fermentation with RP and BP (p > 0.05); however, the distribution tended to be scattered by RP and BP treatment (Figure 3D). In addition, the S3-Bifidobacterium cluster exhibited a different direction in gut microbiota changes compared to the other samples.
Figure 3.
Analysis of α and β diversity by enterotype following RP and BP fermentation. (A) S1-Phocaeicloa, (B) S2-Prevotella, (C) S3-Bifidobacterium, and (D) non-metric multidimensional scaling (NMDS). CTL, negative control (without fermentable substrate); RP, red beet powder; BP, betanin pigment. * p < 0.05, ** p < 0.01.
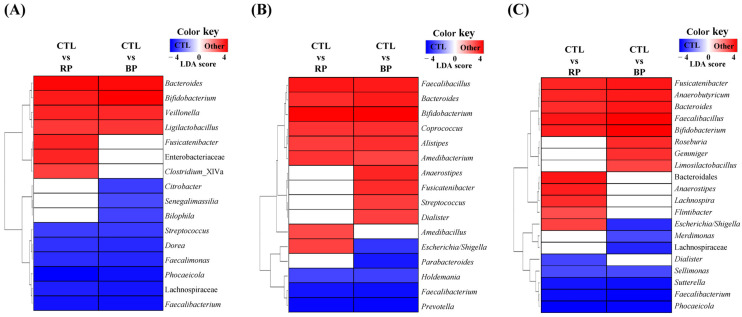
Changes in the microbial composition were assessed to determine the effects of RP and BP on the gut microbiota of each sample. The major phyla Firmicutes and Bacteroidetes did not show significant changes, whereas Actinobacteria and Proteobacteria exhibited significant alterations (p < 0.05; Table S2). At the genus level, the RP and BP treatments significantly changed the abundance of gut microbes (Figure 4). In S1-Phocaeicola, a total of 16 bacteria were significantly altered by RP and BP, with Bacteroides, Bifidobacterium, Veillonella, and Ligilactobacillus increasing in both, while Streptococcus, Dorea, Faecalimonas, Phocaeicola, Lachnospiraceae, and Faecalibacterium decreased (p < 0.05). Similarly, in S2-Prevotella, a total of 16 bacteria were significantly altered by RP and BP, with Faecalibacillus, Bacteroides, Bifidobacterium, Coprococcus, Alistipes, and Amedibacterium increasing in both, whereas Holdemania, Faecalibacterium, and Prevotella decreased (p < 0.05). In the case of S3-Bfidobacterium, a total of 20 bacteria were significantly changed by RP and BP, with Fusicatenibacter, Anaerobutyricum, Bacteroides, Faecalibacillus, and Bifidobacterium increasing in both, whereas Sellimonas, Sutterella, Faecalibacterium, and Phocaeicola decreased (p < 0.05). Among the bacteria that were increased by RP and BP, Bacteroides and Bifidobacterium increased in all subjects, whereas Faecalibacterium decreased. However, the rate of increase in Bifidobacterium was the lowest in the S2-Prevotella sample and the highest in the S3-Bifidobacterium sample (Table S3). In contrast, Phocaeicola, which was the core microbiota of the enterotype, was significantly reduced in the S1-Phocaeicola and S3-Bifidobacterium samples, whereas Prevotella was only present in the S2-Prevotella sample and was reduced.
Figure 4.
Significantly different relative abundance at the genus level by enterotype following RP and BP fermentation. (A) S1-Phocaeicloa, (B) S2-Prevotella, and (C) S3-Bifidobacterium. Significantly different relative abundance was examined using the liner discriminant analysis (LDA) effect size (LEfSe) at the genus level (p < 0.05, LDA score > 3). CTL, negative control (without fermentable substrate); RP, red beet powder; BP, betanin pigment.
3.4. Enterotype-Specific Effects of RP and BP on SCFA Production
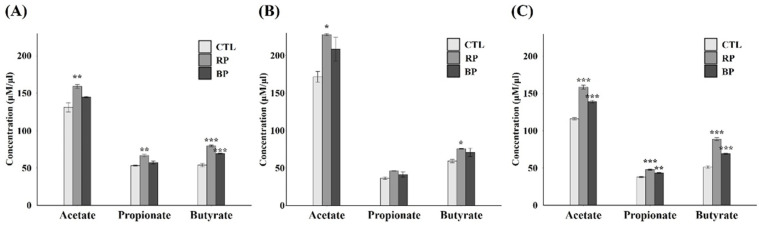
Figure 5 shows the difference in SCFA production between the subjects after fermentation. Acetate and propionate in the S1-Phocaeicola samples significantly increased in RP (p < 0.05), whereas butyrate increased in both RP and BP (p < 0.05). In the S2-Prevotella sample, fermentation with RP significantly increased the acetate and butyrate levels (p < 0.05), whereas no change was observed after fermentation with BP (p > 0.05). In contrast, the S3-Bifidobacterium sample showed significant increases in acetate, propionate, and butyrate levels in both RP and BP groups (p < 0.05).
Figure 5.
Amount of SCFAs by enterotype following RP and BP fermentation. (A) S1-Phocaeicloa, (B) S2-Prevotella, and (C) S3-Bifidobacterium. CTL, negative control (without fermentable substrate); RP, red beet powder; BP, betanin pigment; SCFA, short-chain fatty acid. * p < 0.05, ** p < 0.01, *** p < 0.001.
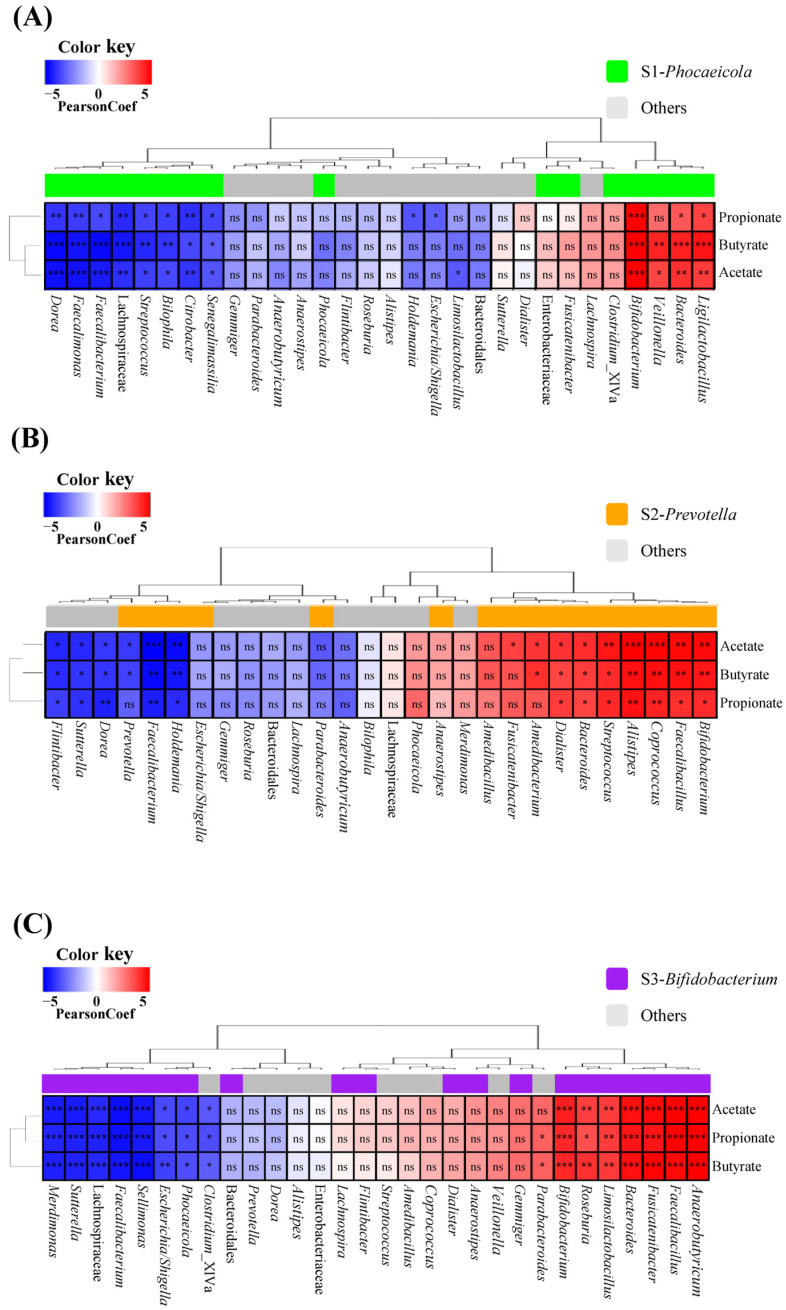
Furthermore, to investigate the association between SCFA production in each subject and their gut microbiota, Pearson correlation analysis was performed between the microbiota and SCFAs that were significantly altered by RP and BP (Figure 6). In general, across all samples, all SCFAs were positively correlated with Bacteroides and Bifidobacterium, whereas they were negatively correlated with Facecalibacterium. Meanwhile, in the S1-Phocaeicola sample, Veillonella and Ligilactobacillus, which were increased by RP and BP, were positively correlated with most SCFAs, whereas Streptococcus, Dorea, Faecalimonas, and Lachnospiraceae, which were decreased by RP and BP, were negatively correlated with all SCFAs. The genera Faecalibacillus, Coprococcus, Alistipes, and Amedibacterium, which were increased by RP and BP in the S2-Prevotella sample, were positively correlated with most SCFAs, while the genera Holdemania and Prevotella, which were decreased by RP and BP, were negatively correlated with SCFAs. For the S3-Bifidobacterium sample, the genera Fusicatenibacter, Anaerobutyricum, and Faecalibacillus, which increased in both the RP and BP, were positively correlated with all SCFAs, whereas Sellimonas, Sutterella, and Phocaeicola, which decreased in both the RP and BP, were negatively correlated. Importantly, certain microorganisms of each enterotype were positively correlated with SCFA production. For example, Ligilactobacillus and Veillonella were only positively correlated with SCFA in the S1-Phocaeicola sample, while Coprococcus and Alistipes were only positively correlated in the S2-Prevotella sample. Furthermore, the genera Anaerobutyrticum and Roseburia positively correlated with all SCFAs in the S3-Bifidobacterium sample.
Figure 6.
Pearson correlation analysis between fecal microbiota and SCFAs by enterotype. (A) S1-Phocaeicloa, (B) S2-Prevotella, and (C) S3-Bifidobacterium. ns, non-significant; SCFA, short-chain fatty acid. * p < 0.05, ** p < 0.01, *** p < 0.001.
3.5. Enterotype-Specific Effects of RP and BP on Microbial Metabolic Activities
Microbial metabolic activities were predicted using PICRUSt2 by comparing the microbiota function against sequencing data and the KEGG database. Figure S4 shows the pathways significantly altered by RP and BP at KEGG Level 2 for each participant. In the S1-Phocaeicola sample, no significant alterations of the pathways were observed, whereas in the S2-Prevotella sample, the metabolism of cofactors and vitamins was consistently reduced. In contrast, in the S3-Bifidobacterium sample, the metabolism of carbohydrates, terpenoids, and polyketides was increased by RP and BP, whereas energy metabolism, cell growth, and cell death were decreased. In the S2-Prevotella sample, the decrease in cofactor and vitamin metabolism was primarily due to riboflavin metabolism. In the S3-Bifidobacterium sample, the increase in carbohydrate metabolism was mainly due to glycolysis/gluconeogenesis and the metabolism of starch, sucrose, pyruvate, galactose, amino sugars, nucleotide sugars, glyoxylates, and dicarboxylates (Figure 7B,C). In contrast with the results at KEGG level 2, at level 3 in the S1-Phocaeicaola sample, methane metabolism and phenylalanine, tyrosine, and tryptophan biosynthesis were significantly changed by RP and BP (Figure 7A).
Figure 7.
Significantly different relative abundance of PICRUSt2-Predicted microbial metabolic activities by enterotype at Level 3 of KEGG pathway following BP and BD fermentation. (A) S1-Phocaeicloa, (B) S2-Prevotella, and (C) S3-Bifidobacterium. Significantly different relative abundance was calculated using Welch’s t-test (p < 0.05, Difference proportions > 0.01). CTL, negative control (without fermentable substrate); RP, red beet powder; BP, betanin pigment.
4. Discussion
In the current study, we aimed to investigate the in vitro enterotype-specific changes in the gut microbiota and SCFAs using RP and BP. We confirmed the hypothesis that the effects of RP and BP on the gut microbiota are enterotype-specific. Specifically, it depended on the relative abundance of Bifidobacterium and Prevotella. The results of the present study emphasize the importance of enterotypes in personalized nutrition.
We first analyzed the gut microbiota of 30 subjects and then selected one subject per enterotype for fecal fermentation. Three enterotypes were identified, and it was confirmed that the subjects selected for each enterotype truly represented their respective enterotypes. The enterotype is commonly divided into Prevotella, Bacteroides, and Ruminococcus types [10]; however, our results showed that it was divided into Phocaeicola, Prevotella, and Bifidobacterium types in our subjects. Lu et al. reported that enterotypes can vary depending on diet, country, regional environment, and other factors [38]. Previous studies have shown that the gut microbiota of Africans is distributed between Prevotella and Bacteroides, whereas Bifidobacterium and Bacteroides are the core microbiota in Asian and Western populations, respectively [12]. This indicates that the enterotype may vary depending on the sample ranges and that people in each region may have an enterotype distribution with corresponding regional characteristics. Therefore, as shown in Figure 1, Phocaeicola, Prevotella, and Bifidobacterium could be representative enterotypes of the core bacteria.
The diverse gut microbiota interact with each other to perform various physiological, metabolic, and immunological functions. Alpha diversity is used to describe the microbial diversity within an ecosystem. In this study, Shannon (evenness, referring to the number or proportion of bacteria within an ecosystem) and Chao (richness, referring to the total number of microbial taxa within an ecosystem) indices were used to evaluate the effects of RP and BP on alpha diversity. After 6 h of fermentation, the Chao index decreased only in S3-Bifidobacterium samples treated with RP and BP. A previous study reported that easily fermentable substrates promote the growth of certain gut microbiota, significantly reducing alpha diversity [39,40]. Furthermore, the NMDS analysis showed that the S3-Bifidobacterium sample had different directions of RP- and BP-induced changes in the gut microbiota compared to other subjects, similar to studies showing that different enterotypes or subjects have different gut microbiota altered by additives [41,42]. Thus, RP and BP induced changes in the gut microbiota depending on the enterotype, indicating that they can affect host health in a short time, especially in the S3-Bifidobacterium sample.
RP and BP affected the abundance of different bacteria in each participant at the genus level, including increases in potentially beneficial bacteria and decreases in potentially harmful bacteria. Ligilactobacillus (formerly known as Lactobacillus) is a classic probiotic [43], and Veillonella helps maintain a healthy gut environment by producing acetate and propionate from lactic acid produced by Lactobacillus species [44]. Conversely, Dorea was positively associated with body weight, waist circumference, and BMI in overweight/obese subjects [45]. The abundances of these three microbes were altered in the S1-Phocaeicola sample. Meanwhile, in the S2-Preovtella sample, Coprococcus and Alistipes increased with RP and BP, whereas Anaerostipes increased only with BP. It has been reported that Coprococcus alleviates colitis by regulating the gut microbiota and immunoglobulin A [46]. Alistipes have been shown to negatively correlate with several inflammatory factors such as LPS and TNF-α [47]. Also, these two bacteria can produce SCFAs [48,49]. In contrast, in the S3-Bifidobacterium sample, Anaerobutyricum, Limosilactobacillus, and Roseburia increased, whereas Sutterella decreased. Sutterella is a pathogen that worsens the immune system and increases susceptibility to intestinal diseases [50], whereas Anaerobutyricum (known as Eubacterium) is a candidate next-generation probiotic that provides energy to the host and interacts with the immune system by producing SCFAs [51,52]. Limosilactobacillus and Roseburia regulate barrier homeostasis and cytokine release by producing SCFAs [53,54]. Overall, RP and BP contributed to an increase in beneficial bacteria and a decrease in pathogenic bacteria; however, the effects were different for each enterotype. In particular, the bacteria that increased in each subject were the dominant bacteria in individuals, and these results confirmed the intestinal-specific response to RP and BP from the perspective of the gut microbiota. Additionally, Bifidobacterium, Bacteroides, Fusicatenibacter and Anaerostipes, which were elevated in more than two subjects, produce SCFAs and affect host health [55,56,57,58].
The same dietary substrate has been shown to result in different levels of SCFA changes among the enterotypes. It has been shown that fecal fermentation of fibers, such as fructooligosaccharides, sorghum bran, and corn arabinoxylan, significantly increases the total SCFAs and propionate in the Prevotella type compared to the Bacteroides type [39]. In addition, a previous study using marine oligosaccharides and polysaccharides, including alginate and carrageenan, revealed that total SCFAs and butyrate levels were higher in the Bacteroides type than in other enterotypes [59,60]. While most studies on responses to dietary substrates have focused on in vitro experiments, in vivo experiments have also shown that dietary intervention with capsaicin caused a significant increase in butyrate levels in the Bacteroides type after intake by the subjects [61]. In this study, there was a tendency to increase SCFAs in all subjects, as well as an increase in SCFA producers and associated bacteria; however, there were differences in the SCFA changes in each subject. The S2-Prevotella sample showed fewer changes in SCFAs compared to the other samples, with no significant change caused by BP, whereas the S3-Bifidobacterium sample showed a significant increase in all SCFAs owing to the RP and BP treatment. Prevotella produces SCFAs through carbohydrate metabolism in the gut [39]. Previous studies have suggested that Prevotella is negatively correlated with Bifidobacterium and that Bifidobacterium growth may be affected by the enterotype [62]. A clinical study in which subjects were administered red beet juice showed increased Bifidobacterium abundance and decreased Prevotella abundance [22]. Furthermore, Bifidobacterium possesses active ingredients involved in enzymatic deglycosylation, which involves the transformation of betanin [21], and betacyanin complexes in the gut of pigs have been shown to increase Bifidobacterium [25]. Here, Bifidobacterium increased in all subjects after RP and BP fecal fermentation, but the relative abundance and ratio of increase were lowest in the S2-Prevotella sample and highest in the S3-Bifidobacterium sample. The findings of previous research and that of the current study suggest that in the Prevotella-dominant enterotype, consumption of a diet rich in betacyanins may not be effective for SCFA production, as it could potentially hinder the growth of Bifidobacterium and lead to a decrease in Prevotella. Conversely, Bifidobacterium-dominant enterotypes are expected to produce higher SCFAs because Bifidobacterium grows more abundantly than other enterotypes. Meanwhile, RP-treated samples showed higher SCFA production than BP-treated samples, which was believed to be the result of the action of various phytochemicals other than betanin contained in RP.
Previous studies have shown that different microbes correlate with SCFAs depending on the enterotype. Fang et al. [63] reported that following in vitro fecal fermentation with Lactobacillus parabuchneri, the Faecalibacterium-enterotype group was positively correlated with SCFAs with Faecalibacterium, Anaerostipes, Coprococcus, and Butyricicoccus, and the Escherichia/Shigella-enterotype group was positively correlated with Streptococcus, Bacteroides, and Sutterella. A study on the fecal fermentation of bacteriocins showed that the positive association between gut microbiota and SCFAs was more pronounced in the Prevotella enterotype compared to other enterotypes [42]. In this study, SCFA-producing bacteria that were increased by RP and BP were positively correlated with SCFAs; however, bacteria that were increased only in certain subjects were not positively correlated with SCFAs in other subjects. For example, Ligilactobacillus and Veillonella, which increased in the S1-Phocaeicola sample, were positively correlated in the S1-Phocaeicola sample, whereas Coprococcus and Alistipes, which increased in the S2-Prevotella sample, were positively correlated in the S2-Prevotella sample. In addition, Anaerobutyricum, which increased in the S3-Bifidobacteirum sample, was positively correlated only in the S3-Bifidobacteirum sample. Therefore, these results suggest that the gut microbiota involved in SCFA production, which was altered by RP and BP, differed according to enterotype.
Microbial metabolic activities were predicted using PICRUSt2. At KEGG level 2, various metabolisms are altered after RP and BP fecal fermentation, confirming that “metabolism” is the main microbial metabolic activity. In particular, carbohydrate metabolism was significantly increased in the S3-Bifidobacterium group. Starch, sucrose metabolism, and glycolysis/gluconeogenesis were increased in both RP and BP, whereas pyruvate and galactose metabolism were increased only by RP and BP, respectively. Dhananjayan et al. reported that oral administration of betanin increased gluconeogenic enzymes [64]. The glycolysis/gluconeogenesis pathway produces pyruvate and oxaloacetate, the precursors of SCFAs, from alpha-d-glucose-6-phosphate, the end product of starch, sucrose, and galactose metabolism [65]. Although mass spectrometry-based microbial metabolome analysis is needed, these results show that the S3-Bifidobacterium sample had useful changes in gut microbial metabolism following RP and BP compared to the other samples. These results indicate that the microbial metabolic activities altered by RP and BP may differ between enterotypes, especially those related to SCFA generation.
This study has some limitations. A key limitation is the small sample size, with only one subject representing each enterotype (Phocaeicola, Prevotella, and Bifidobacterium). This restricts the generalizability of our findings and reduces statistical power. Additionally, there is potential variability in responses among different populations, which could further impact the applicability of our results. Future research should involve larger and more diverse cohorts to better capture these differences. Another important consideration is that our study relied on in vitro fecal fermentation models, which cannot fully replicate the complexities of gut microbiota interactions in vivo. While useful for examining specific responses, these models do not account for factors such as host immune interactions and gastrointestinal dynamics. Furthermore, although it has been reported that gut microbiota can bioconvert red beetroot and betanin, our study did not confirm this bioconversion. Therefore, follow-up in vivo studies and mass-spectrometry-based catabolite and microbial metabolomics analyses are essential to gain a more comprehensive understanding of the effects of RP and BP on gut health according to enterotype.
Nevertheless, these results provide information on the enterotype-specific effects of RP and BP in modulating gut microbiota and SCFA production. To our knowledge, this is a pioneering in vitro study investigating enterotype-specific changes in gut microbiota and SCFA production in response to RP and BP. By linking these alterations to variations in microbial composition, our findings enhance the understanding of personalized nutrition strategies aimed at optimizing gut health through tailored dietary interventions. This research underscores the importance of enterotype differentiation in the development of functional foods targeting gut microbiota and SCFA production. Additionally, as we consider the implications of enterotype-specific responses to RP and BP, the interplay between host health and gut microbiota underscores the need for personalized medicine, as broad interventions often lack specificity. Advancements in nanocarriers could enhance targeted delivery, enabling the co-encapsulation of probiotics and prebiotics for more effective, tailored treatments [66].
5. Conclusions
Thirty Korean subject enterotypes were divided into Phocaeicola, Prevotella, and Bifidobacterium enterotypes, and the effects of RP and BP as prebiotics on the gut microbiota differed depending on the enterotype. The S3-Bifidobacterium sample showed a significant decrease in the Chao index and a different trend in gut microbiota changes in the NMDS compared to the other samples. The increase in SCFAs was greatest in the S3-Bifidobacterium sample, whereas the S2-Prevotella sample showed a smaller change in SCFAs. In addition, our results showed that bacterial and microbial metabolic activities related to SCFAs were different for each enterotype. These results indicate that RP and BP have enterotype-specific responses in the gut microbiota and SCFA production.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life14111391/s1, Figure S1: Taxonomy composition of 30 subjects at the phylum level. The *, #, and & indicated significantly different abundance in each group (*; Phocaeicola type, #; Prevotella type, &; Bifidobacterium type). Significant different relative abundance analysis was examined using LEfSe (p < 0.05, LDA score > 3); Figure S2: Tree analysis for gut microbiota clustering of 30 subjects and the selected subjects; Figure S3: Taxonomy composition of selected subjects at the phylum level. The *, #, and & indicated significantly different abundance in each group (*; S1-Phocaeicola, #; S2-Prevotella, &; S3-Bifidobacterium). Significant different relative abundance analysis was examined using LEfSe (p < 0.05, LDA score > 3); Figure S4: Effects of RP and BP on Predicted microbial metabolic activities by enterotype at Level 2 of KEGG pathway (p < 0.01). (A) S1-Phocaeicola, (B) S2-Prevotella and (C) S3-Bifidobacterium. BLK, negative control (without fermentable substrate); RP, red beet powder; BP, betanin pigment; Table S1: General characteristics of subjects according to the enterotypes; Table S2: Taxonomy composition at the phylum level after fecal fermentation; Table S3: Relative abundance of Phocaeicola, Prevotella, and Bifidobacterium after fecal fermentation.
Author Contributions
Conceptualization, G.-P.K., J.K., K.-H.B. and C.S.K.; methodology, G.-P.K., J.K., K.-H.B. and C.S.K.; software, G.-P.K., H.J. and J.K.; validation, G.-P.K., K.-H.B. and C.S.K.; formal analysis, G.-P.K.; investigation, G.-P.K. and H.J.; resources, J.S.K.; data curation, G.-P.K. and H.J.; writing—original draft preparation, G.-P.K.; writing—review and editing, G.-P.K. and C.S.K.; visualization, G.-P.K., H.J. and J.K.; supervision, C.S.K.; project administration, C.S.K.; funding acquisition J.S.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Jeju National University (approval number: JJNU-IRB-2024-026; approval date: 19 March 2024). Also, this trial was registered with the Clinical Research Information Service as KCT0009657.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data generated in this study are presented in this paper. Further details are available upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ0161732023), the Rural Development Administration, Republic of Korea, and a Grant (No. 2016R1A6A1A03012862) from the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zhu B., Wang X., Li L. Human gut microbiome: The second genome of human body. Protein Cell. 2010;1:718–725. doi: 10.1007/s13238-010-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomaa E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek. 2020;113:2019–2040. doi: 10.1007/s10482-020-01474-7. [DOI] [PubMed] [Google Scholar]
- 3.Barandouzi Z.A., Starkweather A.R., Henderson W.A., Gyamfi A., Cong X.S. Altered composition of gut microbiota in depression: A systematic review. Front. Psychiatry. 2020;11:536093. doi: 10.3389/fpsyt.2020.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penders J., Stobberingh E.E., Brandt P.v.d., Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 5.Jose P.A., Raj D. Gut microbiota in hypertension. Curr. Opin. Nephrol. Hypertens. 2015;24:403–409. doi: 10.1097/MNH.0000000000000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramakrishna B.S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 2013;28:9–17. doi: 10.1111/jgh.12294. [DOI] [PubMed] [Google Scholar]
- 7.Liu P., Wang Y., Yang G., Zhang Q., Meng L., Xin Y., Jiang X. The role of short-chain fatty acids in intestinal barrier function, inflammation, oxidative stress, and colonic carcinogenesis. Pharmacol. Res. 2021;165:105420. doi: 10.1016/j.phrs.2021.105420. [DOI] [PubMed] [Google Scholar]
- 8.Wu Z., Tian E., Chen Y., Dong Z., Peng Q. Gut microbiota and its roles in the pathogenesis and therapy of endocrine system diseases. Microbiol. Res. 2023;268:127291. doi: 10.1016/j.micres.2022.127291. [DOI] [PubMed] [Google Scholar]
- 9.Portincasa P., Bonfrate L., Vacca M., De Angelis M., Farella I., Lanza E., Khalil M., Wang D.Q.-H., Sperandio M., Di Ciaula A. Gut microbiota and short chain fatty acids: Implications in glucose homeostasis. Int. J. Mol. Sci. 2022;23:1105. doi: 10.3390/ijms23031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.-M. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.H., Lee H., You H.S., Sung H.-J., Hyun S.H. Metabolic pathway prediction of core microbiome based on enterotype and orotype. Front. Cell. Infect. Microbiol. 2023;13:1173085. doi: 10.3389/fcimb.2023.1173085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mobeen F., Sharma V., Tulika P. Enterotype variations of the healthy human gut microbiome in different geographical regions. Bioinformation. 2018;14:560. doi: 10.6026/97320630014560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartsch M., Hahn A., Berkemeyer S. Bridging the gap from enterotypes to personalized dietary recommendations: A metabolomics perspective on microbiome research. Metabolites. 2023;13:1182. doi: 10.3390/metabo13121182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon M.C., Sina C., Ferrario P.G., Daniel H., Working Group “Personalized Nutrition” of the German Nutrition Society Gut microbiome analysis for personalized nutrition: The state of science. Mol. Nutr. Food Res. 2023;67:2200476. doi: 10.1002/mnfr.202200476. [DOI] [PubMed] [Google Scholar]
- 15.EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS) Scientific opinion on the re-evaluation of beetroot red (e 162) as a food additive. EFSA J. 2015;13:4318. doi: 10.2903/j.efsa.2015.4318. [DOI] [Google Scholar]
- 16.Calogero G., Bartolotta A., Di Marco G., Di Carlo A., Bonaccorso F. Vegetable-based dye-sensitized solar cells. Chem. Soc. Rev. 2015;44:3244–3294. doi: 10.1039/C4CS00309H. [DOI] [PubMed] [Google Scholar]
- 17.Li Q., Shen Y., Guo X., Xu Y., Mao Y., Wu Y., He F., Wang C., Chen Y., Yang Y. Betanin Dose-Dependently Ameliorates Allergic Airway Inflammation by Attenuating Th2 Response and Upregulating cAMP–PKA–CREB Pathway in Asthmatic Mice. J. Agric. Food Chem. 2022;70:3708–3718. doi: 10.1021/acs.jafc.2c00205. [DOI] [PubMed] [Google Scholar]
- 18.de Oliveira S.P.A., do Nascimento H.M.A., Sampaio K.B., de Souza E.L. A review on bioactive compounds of beet (Beta vulgaris L. subsp. vulgaris) with special emphasis on their beneficial effects on gut microbiota and gastrointestinal health. Crit. Rev. Food Sci. Nutr. 2021;61:2022–2033. doi: 10.1080/10408398.2020.1768510. [DOI] [PubMed] [Google Scholar]
- 19.Inoue H., Takayama K., Takahara C., Tabuchi N., Okamura N., Narahara N., Kojima E., Date Y., Tsuruta Y. Determination of Short-Chain Fatty Acids in Mouse Feces by High-Performance Liquid Chromatography Using 2-Nitrophenylhydrazine as a Labeling Reagent. Biol. Pharm. Bull. 2019;42:845–849. doi: 10.1248/bpb.b18-01017. [DOI] [PubMed] [Google Scholar]
- 20.Luo B., Wen Y., Ye F., Wu Y., Li N., Farid M.S., Chen Z., El-Seedi H.R., Zhao C. Bioactive Phytochemicals and Their Potential Roles in Modulating Gut Microbiota. J. Agric. Food Res. 2023;12:100583. doi: 10.1016/j.jafr.2023.100583. [DOI] [Google Scholar]
- 21.Daliri E.B.-M., Baltriukienė D., Burokas A. Beetroot for managing diabetes and its associated gut dysbiosis: Current findings and challenges. Trends Food Sci. Technol. 2023;142:104216. doi: 10.1016/j.tifs.2023.104216. [DOI] [Google Scholar]
- 22.Wang Y., Do T., Marshall L.J., Boesch C. Effect of two-week red beetroot juice consumption on modulation of gut microbiota in healthy human volunteers—A pilot study. Food Chem. 2023;406:134989. doi: 10.1016/j.foodchem.2022.134989. [DOI] [PubMed] [Google Scholar]
- 23.Capper T.E., Houghton D., Stewart C.J., Blain A.P., McMahon N., Siervo M., West D.J., Stevenson E.J. Whole beetroot consumption reduces systolic blood pressure and modulates diversity and composition of the gut microbiota in older participants. NFS J. 2020;21:28–37. doi: 10.1016/j.nfs.2020.08.001. [DOI] [Google Scholar]
- 24.He J., Liu X., Zhang J., Wang R., Cao X., Liu G. Gut microbiome-derived hydrolases—An underrated target of natural product metabolism. Front. Cell. Infect. Microbiol. 2024;14:1392249. doi: 10.3389/fcimb.2024.1392249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Adekolurejo O.O., Wang B., McDermott K., Do T., Marshall L.J., Boesch C. Bioavailability and excretion profile of betacyanins–variability and correlations between different excretion routes. Food Chem. 2024;437:137663. doi: 10.1016/j.foodchem.2023.137663. [DOI] [PubMed] [Google Scholar]
- 26.Brodkorb A., Egger L., Alminger M., Alvito P., Assunção R., Ballance S., Bohn T., Bourlieu-Lacanal C., Boutrou R., Carrière F. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019;14:991–1014. doi: 10.1038/s41596-018-0119-1. [DOI] [PubMed] [Google Scholar]
- 27.Li L., Abou-Samra E., Ning Z., Zhang X., Mayne J., Wang J., Cheng K., Walker K., Stintzi A., Figeys D. An in vitro model maintaining taxon-specific functional activities of the gut microbiome. Nat. Commun. 2019;10:4146. doi: 10.1038/s41467-019-12087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The silva ribosomal rna gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. Vsearch: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole J.R., Chai B., Farris R.J., Wang Q., Kulam-Syed-Mohideen A., McGarrell D.M., Bandela A., Cardenas E., Garrity G.M., Tiedje J.M. The ribosomal database project (RDP-II): Introducing myrdp space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westcott S.L., Schloss P.D. Opticlust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere. 2017;2:e00073-17. doi: 10.1128/mSphereDirect.00073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C., Mai J., Cao X., Burberry A., Cominelli F., Zhang L. Ggpicrust2: An r package for picrust2 predicted functional profile analysis and visualization. Bioinformatics. 2023;39:btad470. doi: 10.1093/bioinformatics/btad470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beals E.W. Advances in Ecological Research. Elsevier; Amsterdam, The Netherlands: 1984. Bray-curtis ordination: An effective strategy for analysis of multivariate ecological data; pp. 1–55. [DOI] [Google Scholar]
- 36.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J., Zhang L., Zhai Q., Zhao J., Zhang H., Lee Y.-K., Lu W., Li M., Chen W. Chinese gut microbiota and its associations with staple food type, ethnicity, and urbanization. Npj Biofilms Microbiomes. 2021;7:71. doi: 10.1038/s41522-021-00245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen T., Long W., Zhang C., Liu S., Zhao L., Hamaker B.R. Fiber-utilizing capacity varies in Prevotella-versus Bacteroides-dominated gut microbiota. Sci. Rep. 2017;7:2594. doi: 10.1038/s41598-017-02995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose D.J., Poudel R., Van Haute M.J., Yang Q., Wang L., Singh M., Liu S. Pulse processing affects gas production by gut bacteria during in vitro fecal fermentation. Food Res. Int. 2021;147:110453. doi: 10.1016/j.foodres.2021.110453. [DOI] [PubMed] [Google Scholar]
- 41.Luo S., Hou Y., Xie L., Zhang H., Liu C., Chen T. Effects of microwave on the potential microbiota modulating effects of agro-industrial by-product fibers among different individuals. LWT. 2023;178:114621. doi: 10.1016/j.lwt.2023.114621. [DOI] [Google Scholar]
- 42.Pu J., Hang S., Liu M., Chen Z., Xiong J., Li Y., Wu H., Zhao X., Liu S., Gu Q. A class iib bacteriocin plantaricin nc8 modulates gut microbiota of different enterotypes in vitro. Front. Nutr. 2022;9:877948. doi: 10.3389/fnut.2022.877948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carbonne C., Chadi S., Kropp C., Molimard L., Chain F., Langella P., Martin R. Ligilactobacillus salivarius cncm i-4866, a potential probiotic candidate, shows anti-inflammatory properties in vitro and in vivo. Front. Microbiol. 2023;14:1270974. doi: 10.3389/fmicb.2023.1270974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N., Wang H., Zhao H., Wang M., Cai J., Hao Y., Yu J., Jiang Y., Lü X., Liu B. Cooperative interactions between Veillonella ratti and Lactobacillus acidophilus ameliorate dss-induced ulcerative colitis in mice. Food Funct. 2023;14:10475–10492. doi: 10.1039/D3FO03898J. [DOI] [PubMed] [Google Scholar]
- 45.Companys J., Gosalbes M.J., Pla-Pagà L., Calderón-Pérez L., Llauradó E., Pedret A., Valls R.M., Jiménez-Hernández N., Sandoval-Ramirez B.A., Del Bas J.M. Gut microbiota profile and its association with clinical variables and dietary intake in overweight/obese and lean subjects: A cross-sectional study. Nutrients. 2021;13:2032. doi: 10.3390/nu13062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang R., Shan S., Shi J., Li H., An N., Li S., Cui K., Guo H., Li Z. Coprococcus eutactus, a potent probiotic, alleviates colitis via acetate-mediated iga response and microbiota restoration. J. Agric. Food Chem. 2023;71:3273–3284. doi: 10.1021/acs.jafc.2c06697. [DOI] [PubMed] [Google Scholar]
- 47.Zhu L., Sha L., Li K., Wang Z., Wang T., Li Y., Liu P., Dong X., Dong Y., Zhang X. Dietary flaxseed oil rich in omega-3 suppresses severity of type 2 diabetes mellitus via anti-inflammation and modulating gut microbiota in rats. Lipids Health Dis. 2020;19:20. doi: 10.1186/s12944-019-1167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Notting F., Pirovano W., Sybesma W., Kort R. The butyrate-producing and spore-forming bacterial genus Coprococcus as a potential biomarker for neurological disorders. Gut Microbiome. 2023;4:e16. doi: 10.1017/gmb.2023.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakamoto M., Ikeyama N., Ogata Y., Suda W., Iino T., Hattori M., Ohkuma M. Alistipes communis sp. Nov., Alistipes dispar sp. Nov. And Alistipes onderdonkii subsp. Vulgaris subsp. Nov., isolated from human faeces, and creation of Alistipes onderdonkii subsp. onderdonkii subsp. Nov. Int. J. Syst. Evol. Microbiol. 2020;70:473–480. doi: 10.1099/ijsem.0.003778. [DOI] [PubMed] [Google Scholar]
- 50.Moon C., Baldridge M.T., Wallace M.A., Burnham C.-A.D., Virgin H.W., Stappenbeck T.S. Vertically transmitted faecal iga levels determine extra-chromosomal phenotypic variation. Nature. 2015;521:90–93. doi: 10.1038/nature14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almeida D., Machado D., Andrade J.C., Mendo S., Gomes A.M., Freitas A.C. Evolving trends in next-generation probiotics: A 5w1h perspective. Crit. Rev. Food Sci. Nutr. 2020;60:1783–1796. doi: 10.1080/10408398.2019.1599812. [DOI] [PubMed] [Google Scholar]
- 52.Engels C., Ruscheweyh H.-J., Beerenwinkel N., Lacroix C., Schwab C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front. Microbiol. 2016;7:184615. doi: 10.3389/fmicb.2016.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abuqwider J., Altamimi M., Mauriello G. Limosilactobacillus reuteri in health and disease. Microorganisms. 2022;10:522. doi: 10.3390/microorganisms10030522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nie K., Ma K., Luo W., Shen Z., Yang Z., Xiao M., Tong T., Yang Y., Wang X. Roseburia intestinalis: A beneficial gut organism from the discoveries in genus and species. Front. Cell. Infect. Microbiol. 2021;11:757718. doi: 10.3389/fcimb.2021.757718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takada T., Kurakawa T., Tsuji H., Nomoto K. Fusicatenibacter saccharivorans gen. Nov., sp. Nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2013;63:3691–3696. doi: 10.1099/ijs.0.045823-0. [DOI] [PubMed] [Google Scholar]
- 56.Schwiertz A., Hold G.L., Duncan S.H., Gruhl B., Collins M.D., Lawson P.A., Flint H.J., Blaut M. Anaerostipes caccae gen. Nov., sp. Nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 2002;25:46–51. doi: 10.1078/0723-2020-00096. [DOI] [PubMed] [Google Scholar]
- 57.Bui T.P.N., Mannerås-Holm L., Puschmann R., Wu H., Troise A.D., Nijsse B., Boeren S., Bäckhed F., Fiedler D., Devos W.M. Conversion of dietary inositol into propionate and acetate by commensal Anaerostipes associates with host health. Nat. Commun. 2021;12:4798. doi: 10.1038/s41467-021-25081-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rivière A., Selak M., Lantin D., Leroy F., De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu T., Pan L., Shang Q., Yu G. Fermentation of alginate and its derivatives by different enterotypes of human gut microbiota: Towards personalized nutrition using enterotype-specific dietary fibers. Int. J. Biol. Macromol. 2021;183:1649–1659. doi: 10.1016/j.ijbiomac.2021.05.135. [DOI] [PubMed] [Google Scholar]
- 60.Fu T., Zhou L., Fu Z., Zhang B., Li Q., Pan L., Zhou C., Zhao Q., Shang Q., Yu G. Enterotype-specific effect of human gut microbiota on the fermentation of marine algae oligosaccharides: A preliminary proof-of-concept in vitro study. Polymers. 2022;14:770. doi: 10.3390/polym14040770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang C., Zhang Y., Zhu X., Liu K., Wang X., Chen M., Wang J., Chen H., Hui S., Huang L. Healthy subjects differentially respond to dietary capsaicin correlating with specific gut enterotypes. J. Clin. Endocrinol. Metab. 2016;101:4681–4689. doi: 10.1210/jc.2016-2786. [DOI] [PubMed] [Google Scholar]
- 62.Chen J., Li Z., Wang X., Fan B., Deng F., Yu H.D., Ze X., Zhu L., Yin Y., Chen Y. Isomaltooligosaccharides sustain the growth of Prevotella both in vitro and in animal models. Microbiol. Spectr. 2022;10:e02621-21. doi: 10.1128/spectrum.02621-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang F., Li Y., Lu X., Wu K., Zhou L., Sun Y., Wu J., Gao J. Effect of potential postbiotics derived from food-isolated Lactobacillus parabuchneri on different enterotypes of human gut microbiome. LWT. 2023;182:114782. doi: 10.1016/j.lwt.2023.114782. [DOI] [Google Scholar]
- 64.Dhananjayan I., Kathiroli S., Subramani S., Veerasamy V. Ameliorating effect of betanin, a natural chromoalkaloid by modulating hepatic carbohydrate metabolic enzyme activities and glycogen content in streptozotocin–nicotinamide induced experimental rats. Biomed. Pharm. 2017;88:1069–1079. doi: 10.1016/j.biopha.2017.01.146. [DOI] [PubMed] [Google Scholar]
- 65.Kingkaw A., Raethong N., Patumcharoenpol P., Suratannon N., Nakphaichit M., Keawsompong S., Roytrakul S., Vongsangnak W. Analyzing predominant bacterial species and potential short-chain fatty acid-associated metabolic routes in human gut microbiome using integrative metagenomics. Biology. 2022;12:21. doi: 10.3390/biology12010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thirumalai A., Girigoswami K., Harini K., Pallavi P., Gowtham P., Girigoswami A. A review of the current state of probiotic nanoencapsulation and its future prospects in biomedical applications. Biocatal. Agric. Biotechnol. 2024;57:103101. doi: 10.1016/j.bcab.2024.103101. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are presented in this paper. Further details are available upon request.