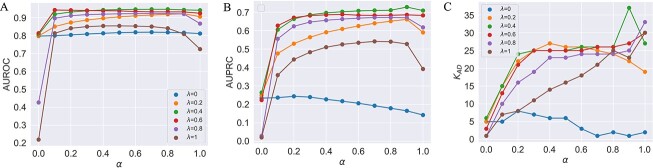
Figure 4.
(A) AUROCs of LIMO-GCN in different parameters (the results are generated by using all negative samples). Comparison of different parameters influencing on LIMO-GCN performance. We systematically varied these parameters to understand their impact and identify the optimal settings for our disease gene prediction task. 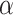 indicates the weight of GCN modules in the model.
indicates the weight of GCN modules in the model. 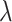 is to balance the loss function by combining L1 loss and cross-entropy loss. (B) AUPRCs of LIMO-GCN in different parameters. (C) No. of AD risk genes (
is to balance the loss function by combining L1 loss and cross-entropy loss. (B) AUPRCs of LIMO-GCN in different parameters. (C) No. of AD risk genes (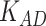 ) in training data among top 100 ranked genes under different parameters.
) in training data among top 100 ranked genes under different parameters.