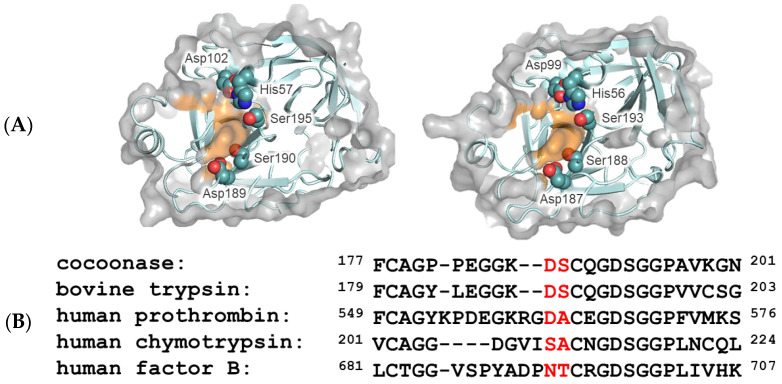
Figure 1.
Molecular modeling of trypsin and cocoonase and their substrate-binding regions. (A) The molecular structures of the substrate-binding regions of trypsin (PDB: 5T3H, left) and Bombyx mori cocoonase (calculated using AlphaFold2, right). Surfaces of the amino acid residues at the S1 site are highlighted in orange. The catalytic triad and substrate-binding residues are indicated. (B) Alignment of the amino acid sequences of the substrate-binding regions of trypsin-like proteases. The predominant substrate-binding residues are indicated by red letters.