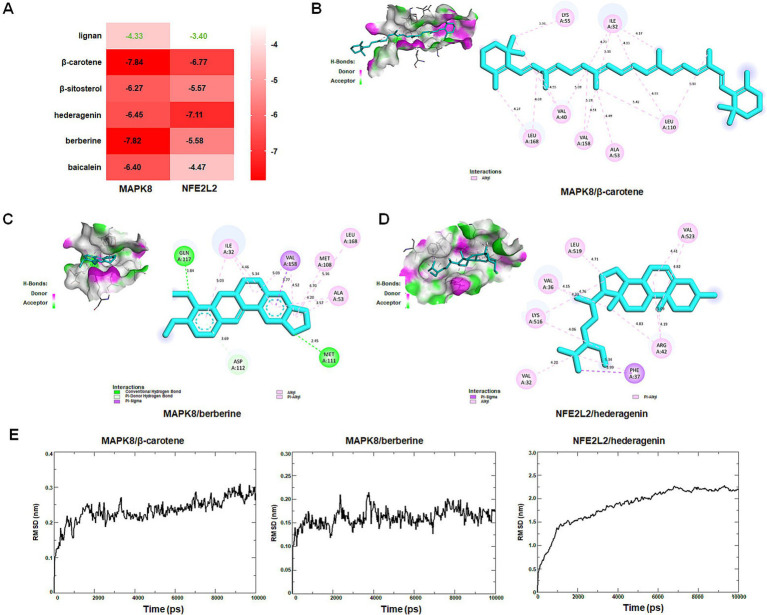
Figure 12.
Identification of new targets by molecular docking and dynamics simulation. (A) The heat map shows the binding free energy (kcal/mol) of molecular docking. (B) Interactions between MAPK8 and β-carotene. (C) Interactions between MAPK8 and berberine. (D) Interactions between NFE2L2 and hederagenin. (E) MAPK8/β-carotene, MAPK8/berberine, and NFE2L2/hederagenin complexes root mean square deviation (RMSD) plot during molecular dynamics simulations. The X-axis represents the time (ps), and the Y-axis represents the RMSD (nm).