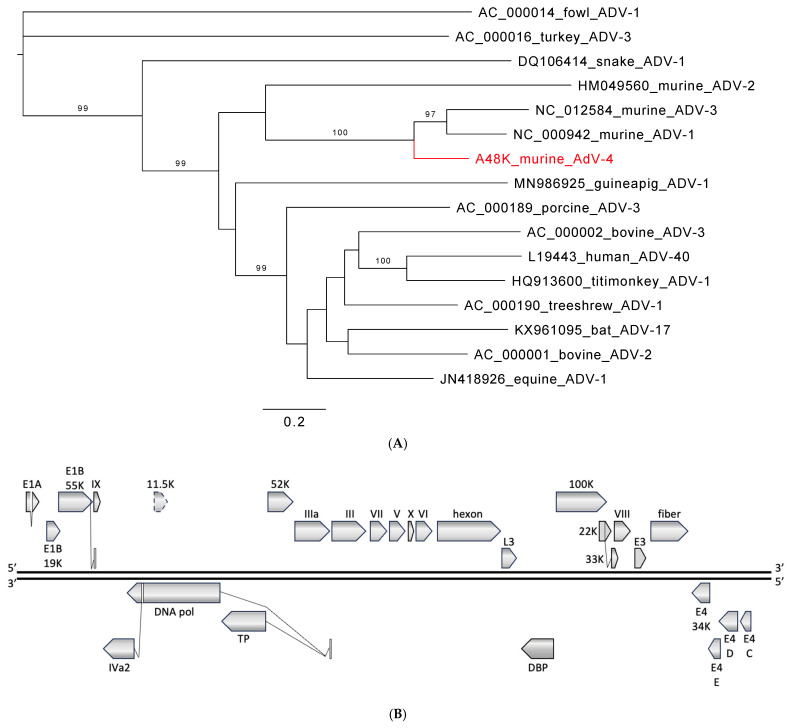
Figure 1.
Murine adenovirus from Zambia. (A) Phylogenetic relationship to selected viruses in the genus Mastadenovirus based on the polymerase amino acid sequence. Phylogeny was reconstructed with the maximum likelihood method applying a Q.pfam+F+I+G4 substitution model as selected by Model Finder implemented in IQtree; bootstrap values (>85%) resulting from 1000 pseudoreplicates are indicated at the respective nodes; the scale bar indicates the number of amino acid substitutions per site, and GenBank accession numbers are given next to the branches. The red font indicates a virus described in this study. (B) Schematic of genome organization. Predicted open reading frames (ORF) and potential splice sites analogous to the other murine adenoviruses are indicated for both strands in all three frames. An ORF with 67% amino acid identity to an 11.5 kDa ORF in the same genome position of HAdV-41 (and other mastadenoviruses) that is not found in the other MAdV is also indicated.