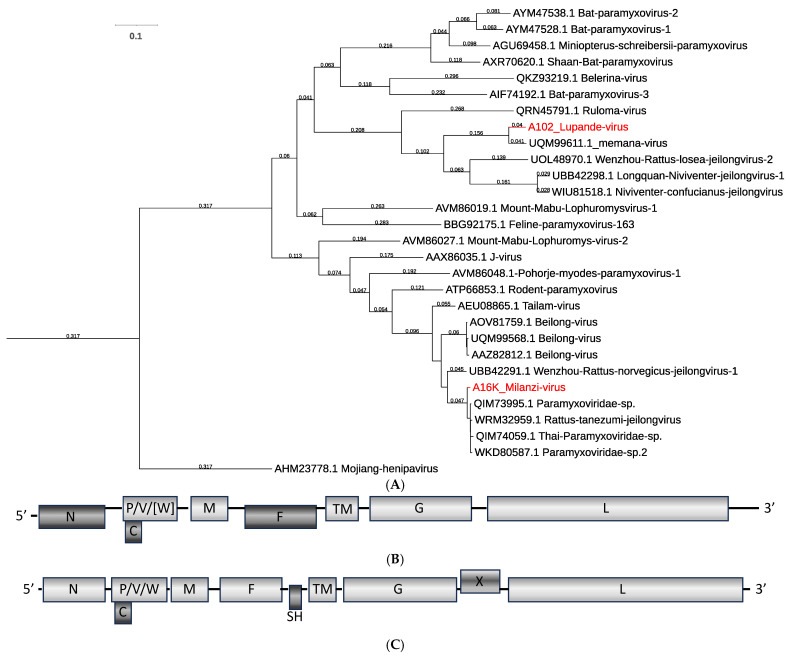
Figure 3.
Jeilongviruses from Zambia. (A) Phylogenetic relationship of Milanzi and Lupande viruses to other viruses in the genus Jeilongvirus based on the polymerase amino acid sequence. Phylogeny was reconstructed using a Clustal W-aligned polymerase amino acid sequence (gap generation and extension penalties of 5 and 1, respectively) with the maximum likelihood method by applying a JTT model as implemented in IQtree; the best bootstrap tree from 500 pseudoreplicates is shown, and branch lengths ≥0.03 substitutions per site are indicated at the respective nodes. GenBank accession numbers are given next to the branches. The red font indicates viruses described in this study. (B) Schematic of Lupande virus genome organization. (C) Schematic of Milanzi virus genome organization. Gray shading indicates the three possible reading frames.