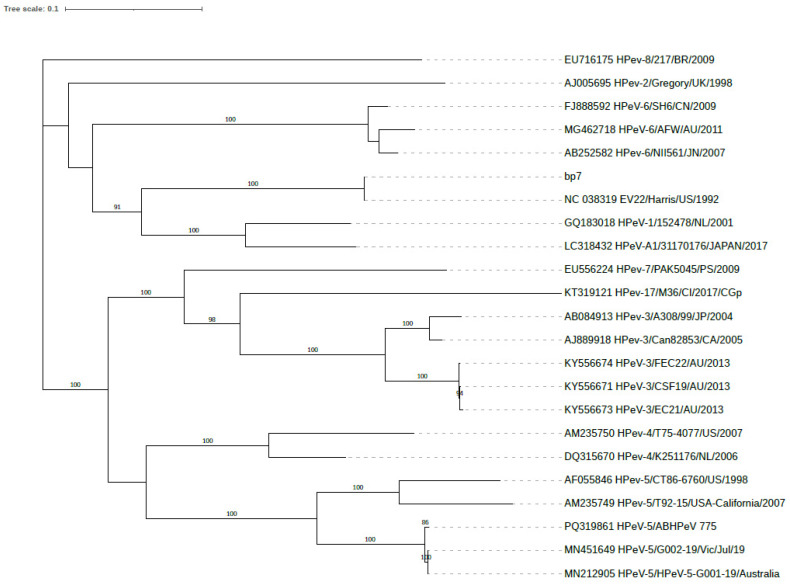
Figure 3.
Maximum likelihood phylogenetic tree by RAxML (GTR + G model) based on HPeV whole-genome sequences. The phylogenetic tree includes a representative HPeV-5-positive sample from Alberta (PQ319861_HPeV-5/ABHPeV_775), a positive control sample HPeV-1_L02971 (bp7), and 21 publicly available HPeV whole-genome sequences from HPeV-genomedb. The Alberta sample (PQ319861_HPeV-5/ABHPeV_775) shares the most recent common ancestor with the recombinant HPeV-5 samples from Australia (13). The positive control sample (bp7-HPeV-1_L02971) is most closely related to other publicly available HPeV-1 virus genomes as expected. The bootstrap values greater than 80 were displayed on the branches. The scale bar corresponds to the expected mean number of nucleotide substitutions per site.