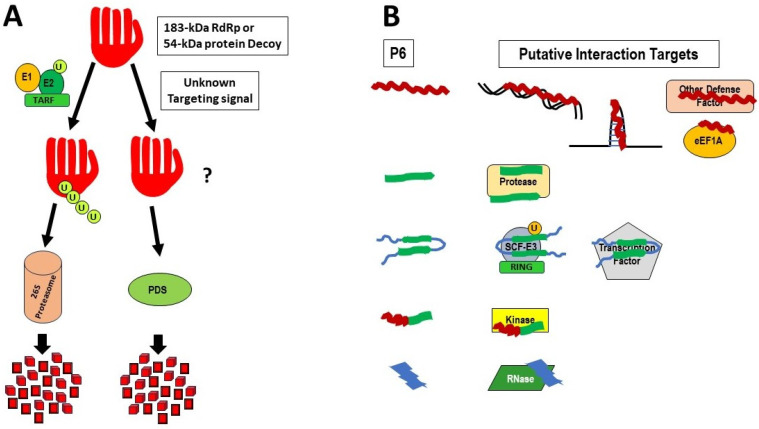
Figure 2.
Models for possible roles of the 54 kDa protein (A) and the P6 proteins (B) during infection: (A) The 54 kDa protein is depicted as a right-handed structure, as described for various RdRps, with the thumb, palm, and fingers representing different substructural regions of the RdRp [85]. As described in the text, the 54 kDa protein may serve as a decoy to prevent the RdRp in the 183 kDa from being degraded, either (left side) by the TARF (TMV-associated RING Finger protein) complex and the 26S proteasome [the ubiquitin-mediated, proteasomal degradation system (UPS)], or (right side) by some other protease-mediated degradation systems (PDS), mediated by unknown targeting signals. TARF is an E3 ubiquitin ligase, acting in concert with ubiquitin-activating enzyme (E1) and ubiquitin-conjugating enzyme (E2), to polyubiquitinate proteins as signals for destruction. (B) The P6 proteins could act as countermeasures to inhibit specific host defense proteins (or interact with other factors, including DNA and RNA) that may have either regulatory or inhibitory roles in the infection cycle of tobamoviruses. The P6 proteins (left side) are shown as representing the various classes of antimicrobial peptides (AMPs), whose structures are classified as α-helices (α; in red); β-sheets [β; in light green, alone or in more complex arrangements including antiparallel β-sheets connected and flanked by random coils (blue)]; combined α-helices and β-sheets (αβ); or non-αβ (blue) [86]. Possible interacting targets include RNA, DNA, other defense factors, eukaryotic elongation factor 1A (eEF1a), proteases, other E3 ubiquitin ligase complexes, transcription factors, kinases, and specific RNases. In the absence of experimental data, the specific associations of particular structures with specific factors are for modeling purposes only.