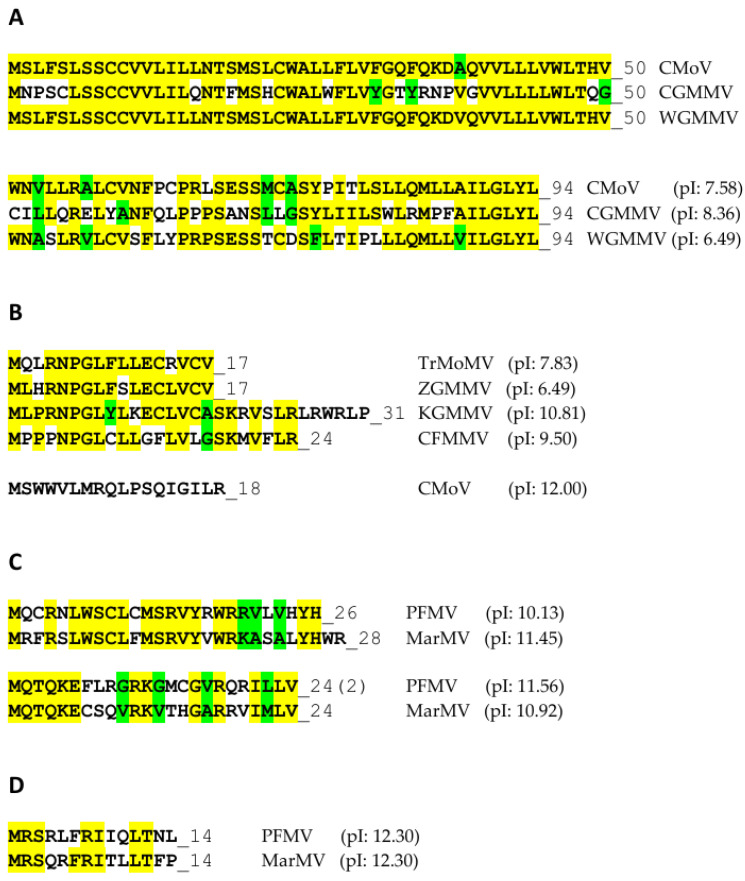
Figure 6.
Conservation of sequences between putative P6 proteins derived from specific non-canonical ORFs 6 of different viruses in the same Subgroup and original host family cluster: (A) The sequence comparisons of Type B, 94-amino acid proteins encoded by three Subgroup 3 Cucurbitaceae-infecting tobamoviruses (CMoV, CGMMV, and WGMMV). (B) The sequence comparisons of Type B, shorter P6 proteins from similar locations in their respective genomes encoded by four Subgroup 2 Cucurbitaceae-infecting tobamoviruses (TrMoMV, ZGMMV, KGMMV, and CFMMV), vs. a similar-sized P6 protein encoded by a different region of CMoV. (C) The sequence comparisons of two sets of Type B, shorter P6 proteins from similar locations in their respective genomes encoded by two Subgroup 4 Passifloraceae-infecting tobamoviruses (PFMV and MarMV). (D) The sequence comparisons of Type D, shorter P6 proteins from similar locations in their respective genomes encoded by PFMV and MarMV. The pIs of the peptide or protein sequences also are shown. Amino acid sequences identical between compared proteins are shown in yellow highlight; those in which the amino acid sequences are similar, are highlighted in light green. Dissimilar amino acids are not highlighted.