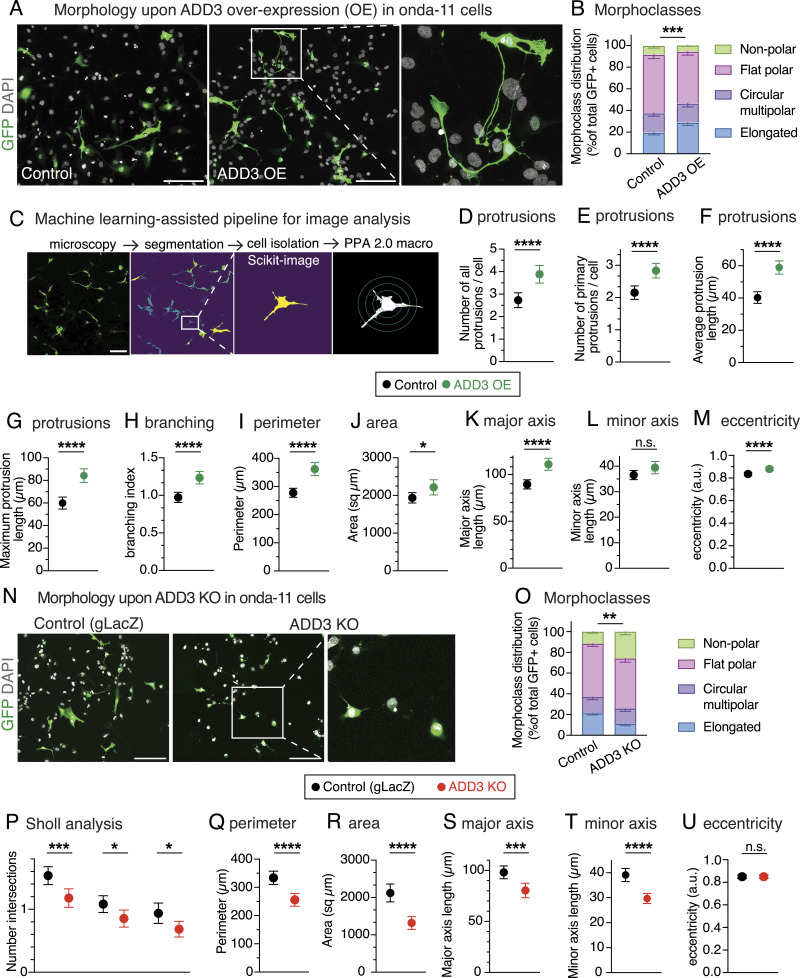
Figure 3. ADD3 regulates Onda-11 glioblastoma stem cell (GSC) morphology and protrusion number.
(A, B, C, D, E, F, G, H, I, J, K, L, M) ADD3 overexpression promotes cell elongation and protrusion abundance. Onda-11 cells were transfected either with GFP and ADD3-overexpressing plasmids (ADD3 OE) or with a GFP and an empty vector (control), and their morphology was analyzed. (A) Representative examples of GFP+ (green) Onda-11 cell morphology in control (left) and ADD3 OE (center). Scale bar: 200 μm. A close-up of elongated cells upon ADD3 OE (right, image width: 250 μm) is shown with the max intensity projection (MIP) of 12 planes. (B) Distribution of the four morphoclasses in control and ADD3 OE Onda-11 GSCs. (C) Schematics of the pipeline for automated cell segmentation and morphological analysis of cells. GFP+ cells from confocal microscopy images (MIPs of 25 planes) are segmented in CellPose, and single cells are isolated to carry out morphological analysis in Python and Fiji using PPA 2.0 macro. Scale bars: 200 μm. Close-up, 177 μm wide. (C, D, E, F, G, H, I, J, K, L, M) Number of total (D) and primary (E) cell protrusions, average (F) and maximum (G) protrusion length, branching index (H), perimeter (I), area (J), major (K) and minor (L) axis length, and eccentricity (M) upon ADD3 OE versus control, calculated as described in (C). (N, O, P, Q, R, S, T, U) ADD3 KO reduces protrusion abundance and induces cell shrinkage. Onda-11 cells were transfected either with an ADD3 KO plasmid or with a gLacZ KO plasmid as a control, and their morphology was analyzed. (N) Representative examples of GFP+ (green) Onda-11 cell morphology in control (left) and ADD3 KO (center). Scale bar: 200 μm. A close-up of cells upon ADD3 KO (right, image width: 300 μm) is shown with the MIP of 12 planes. (O) Distribution of the four morphoclasses in control and ADD3 KO Onda-11 GSCs. (C, P, Q, R, S, T, U) Sholl analysis (P), perimeter (Q), area (R), major (S) and minor (T) axis length, and eccentricity (U) upon ADD3 KO versus control, calculated as described in (C). (B, D, E, F, G, H, I, J, K, L, M, O, P, Q, R, S, T, U) Data are the mean of four (B, D, E, F, G, H, I, J, K, L, M) and eight (O, P, Q, R, S, T, U) independent transfections. (D, E, F, G, H, I, J, K, L, M, P, Q, R, S, T, U) Total number of cells scored: 328 (ADD3 OE) and 397 (control) (D, E, F, G, H, I, J, K, L, M); 317 (KO and control) (P, Q, R, S, T, U). (B, D, E, F, G, H, I, J, K, L, M, O, P, Q, R, S, T, U) Error bars, SEM (B, O), 95% CI (D, E, F, G, H, I, J, K, L, M, P, Q, R, S, T, U); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not statistically significant; two-way ANOVA with Sidak’s post hoc tests (B, O), and t test (D, E, F, G, H, I, J, K, L, M, P, Q, R, S, T, U).