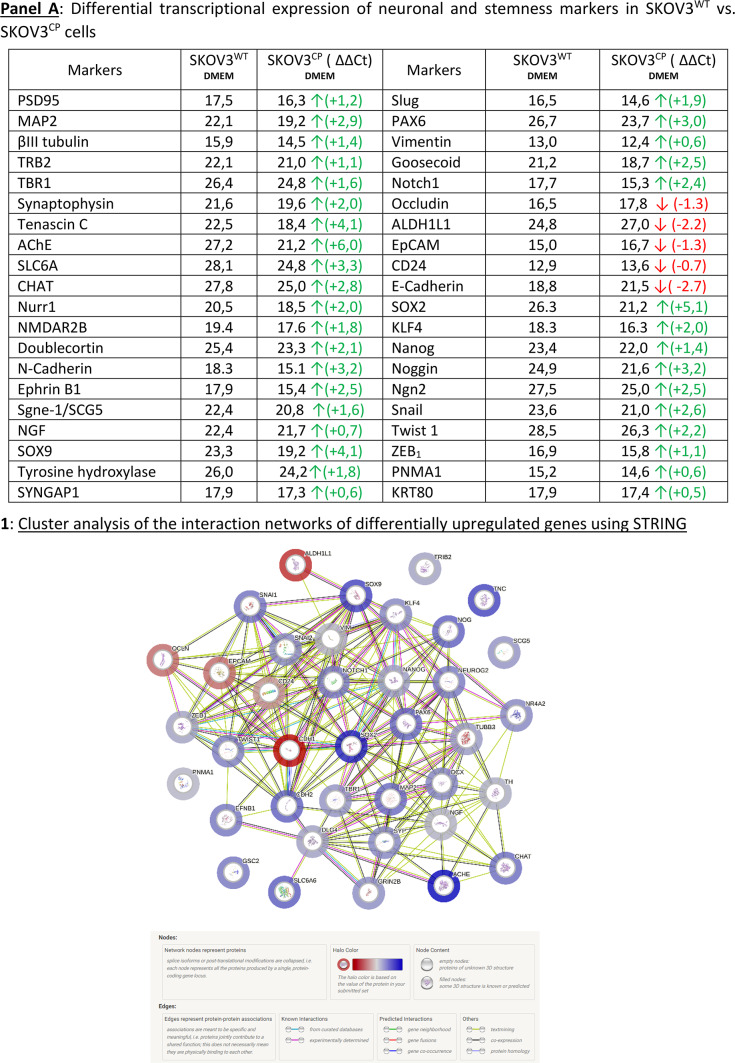
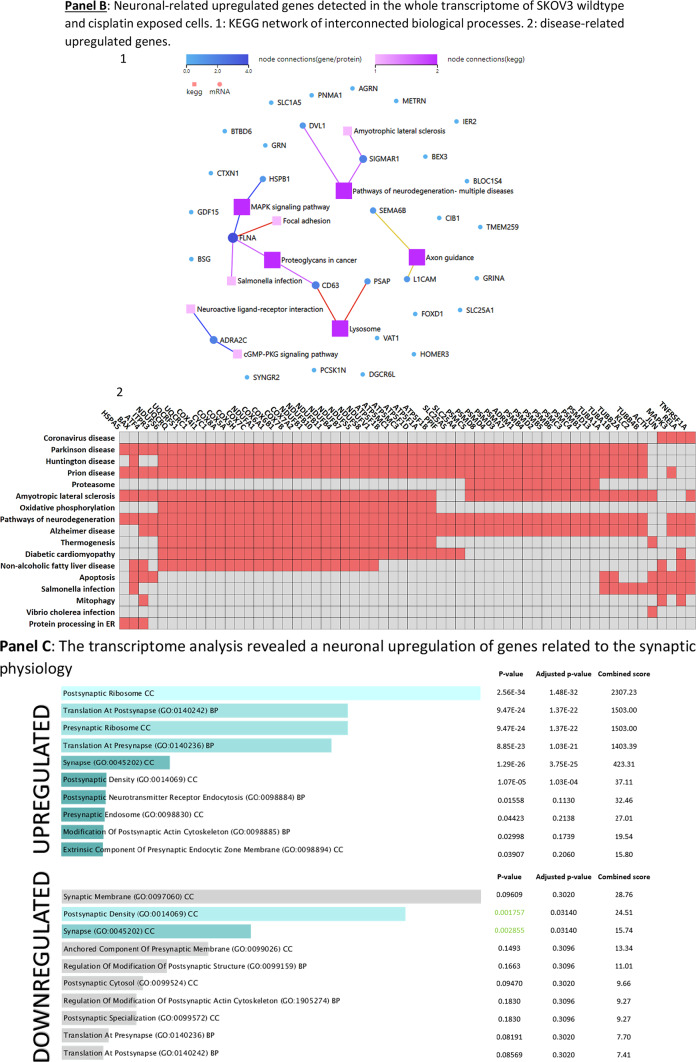
Fig. 4.
Expression of EMT-related proteins in SKOV3CP. Panel A: ΔΔCt values for transcription factors and neuronal markers (RT-qPCR TaqMan system). Transcriptional signatures defining a neuronal phenotype are clearly demonstrated in SKOV3CP cells. Of note, epithelial markers like Occludin, ALDH1L1, EpCAM, CD24 and E-cadherin were significantly downregulated. Green arrows denote an upregulation, red arrows a downregulation at the transcriptional level. Data represents only those genes which were differentially expressed below the ΔCt threshold of 29 cycles. A1: Cluster analysis of the interaction networks of differentially expressed genes using STRING (http://www.string-db.org) with default parameters, including a minimum interaction score of 0.4 (medium confidence) and various interaction sources including Text mining, Experiments, Databases, Co-expression, Neighborhood, Gene Fusion, and Co-occurrence. The colored “halo” around the bubbles indicates differential expression values, with red indicating higher values in SKOV3WT and blue indicating higher values in SKOV3CP. Panel B: 1, Transcriptome analysis of neuronal-related differentially expressed genes in SKOV3WT vs. SKOV3CP cells using NGS. SKOV3 cells were exposed to cisplatin for 72 h to study the first biological response to this agent. We extracted from the transcriptome results the neuronal-related genes found enhanced in SKOV3 cells treated with cisplatin for 72 h and depicted the existence or until unknown interconnection of more than 32 different genes related to neuronal processes found upregulated in cisplatin exposured cells. One relevant gene, which has not escaped our attention, was PNMA1 that codes the paraneoplastic Ma1 antigen a truly specific neuronal protein. 2, Gene expression clustering analysis using the SynGO 2022 database (https://maayanlab.cloud/Enrichr/) revealing the upregulation of several genes related to neurodegenerative diseases and neuronal physiology. Panel C: Gene expression analysis using the SynGO 2022 database from the Enrichr online platform revealed that there are significant enrichments of gene sets associated with synaptic function.